- About
-
Services
-
Offerings
- Offerings
- ADME and Bioanalytical Sciences
- Analytical Chemistry
- Assay Development
- Biochemical Assays
- Biophysical Assays
- Cell Based Assays
- Computational Chemistry
- Fragment and Compound Screening
- Integrated Drug Discovery Services
- Medicinal Chemistry
- Project Management and Consultancy Services
- Protein Expression and Purification Services
- Structural Biology
- Synthetic Chemistry
- Virtual screening
-
Research Phases
- Research Phases
- Hit Identification
- Hit to Lead
- Lead Optimisation
- Therapeutic Areas
- Target Classes
-
Approaches & Techniques
- Approaches & Techniques
- CDH (Target Gene Fragmentation)
- Cryogenic Electron Microscopy (Cryo-EM)
- Differential Scanning Fluorimetry (DSF) and nanoDSF Services
- Direct-to-Biology (D2B)
- Dynamic Light Scattering (DLS)
- eProtein Discovery
- Flow Cytometry
- Fragment Based Drug Discovery (FBDD)
- FragmentBuilder
- Grating-Coupled Interferometry
- High Throughput Screening
- Isothermal Titration Calorimetry (ITC)
- LeadBuilder
- Photoaffinity Labelling
- PoLiPa (Membrane Protein Solubilisation)
- Spectral Shift and MST Services
- Structure Based Drug Design (SBDD)
- Surface Plasmon Resonance (SPR)
- X-ray Crystallography
-
Offerings
- Library
- News & Events
- Careers
CDH Case Studies
A kinase target, a methyltransferase and a DUB target
Our patented Combinatorial Domain Hunting (CDH) technology quickly identifies soluble, highly expressible constructs of target proteins by combining random gene fragmentation with efficient screening of the resulting protein fragments.
CDH is a well-established technique at Domainex, and has been applied to over 60 targets covering a wide range of target classes. For more information about the CDH process, download our brochure here. Three case studies that illustrate the approach are provided below.
Case Study 1: Novel X-Ray Constructs for MEK-1 via CDH
Mitogen-activated protein kinase kinase (MAPKK, also known as MEK-1) is a protein that is over-active in many human cancers.
Challenge
UCB Pharma wished to study the structure of MEK-1 in order to inform its drug discovery programme; however, the protein had previously proved difficult to express and crystallise in their hands.
Solution
Domainex used its proprietary CDH technology on full-length MEK-1 and also on a construct that we designed, in which the disordered region αα 264-307 was replaced with a GSGSGS linker sequence (shown as hatched in Figure 1). The CDH experiment identified 15 soluble, well-expressing fragments of MEK-1 (see Figure 1).
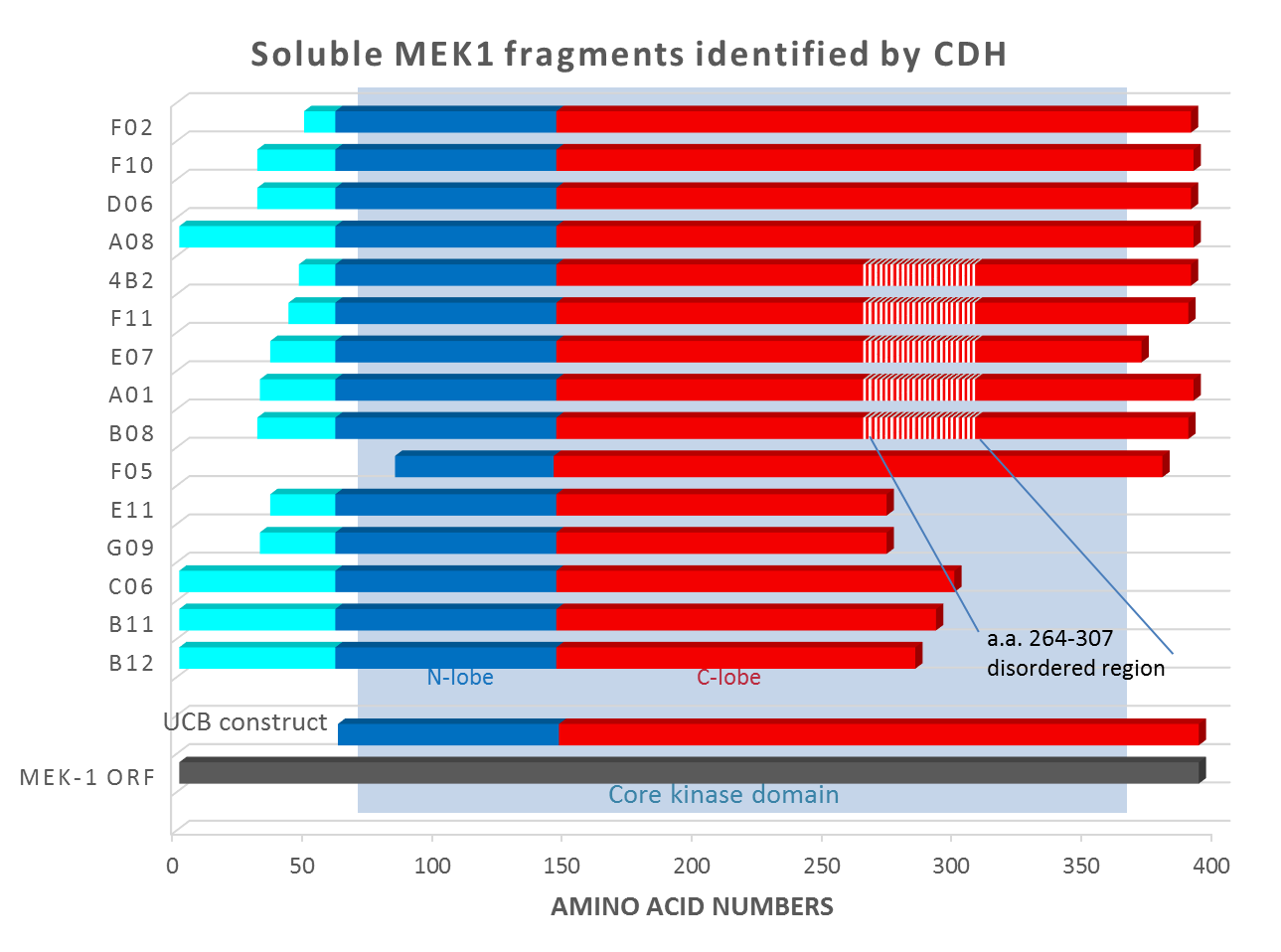
It can be seen that all of these are significantly different from both the full-length protein and the ‘catalytic domain’ construct tried by UCB. The hits were scaled up and subjected to crystallisation trials. Clone F11, which expressed at >10mg/L in E. coli and at >50mg/L in insect cells, was successfully crystallised and gave a high-resolution crystal structure (2.3Å). Further structures were solved for co-crystals with a series of inhibitors: in total, ten inhibitor compounds (described in Laing et al.1) were tested of which nine produced crystals (example shown in Figure 2). Interestingly, for most compounds, diffraction-quality crystals were obtained directly from primary crystallisation screening, without any need to optimise the crystallisation conditions. This indicates that the F11 fragment was readily crystallisable.
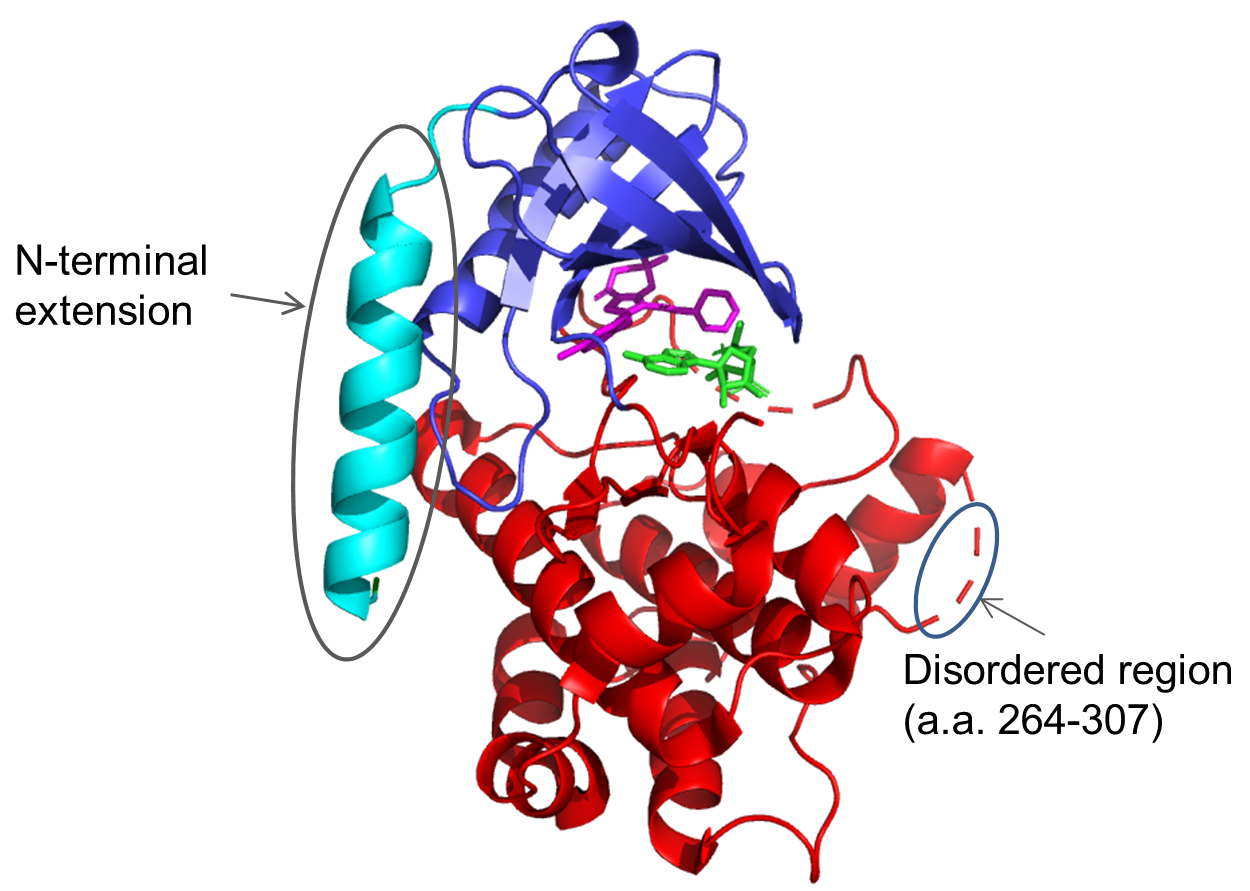
Conclusions
Our CDH technology allowed high-resolution structural information to be obtained for a previously challenging protein. Using this invaluable information, UCB scientists were able to progress their drug discovery programme and design a novel class of molecules which inhibit MEK-1.
This work resulted in a publication.2
Case study 2: USP28 Protein Production via CDH
USP28 is a deubiquitinating enzyme that functions as a regulator of DNA damage response and transcription via c-Myc. Pathophysiologically, USP28 drives colorectal and non-small-cell lung cancer.
Challenge
The Domainex team was asked to identify soluble, stable and highly expressible constructs containing the ubiquitin hydrolase domain. Previous attempts using a bioinformatics approach had produced low expression yields (0.1–0.9mg/L) in E. coli and no structure could be obtained with these clones.
Solution
Domainex used its proprietary CDH technology to screen a library of 52,000 variants of the USP28 sequence. From this, 31 unique, well-expressed constructs were identified, of which 17 covered all or most of the catalytic domain (Figure 3).
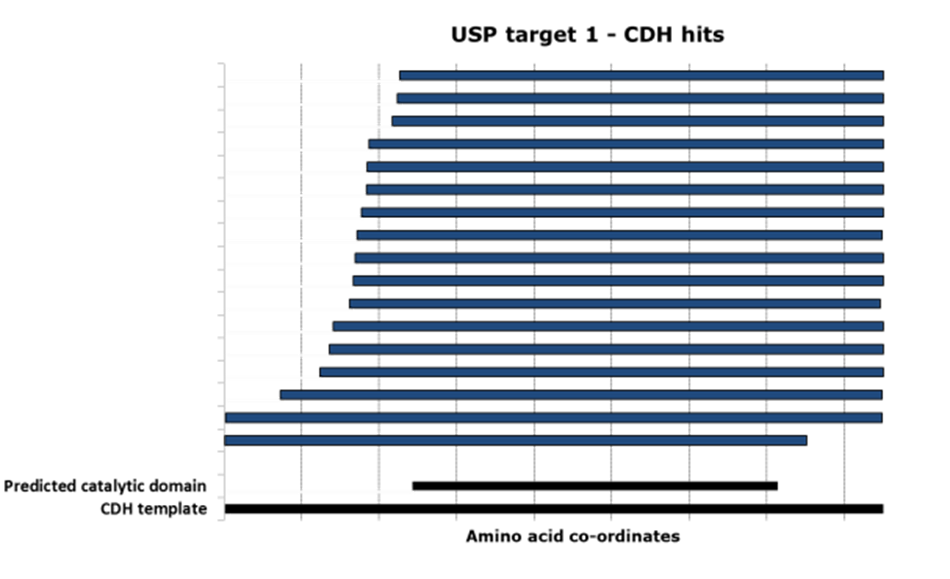
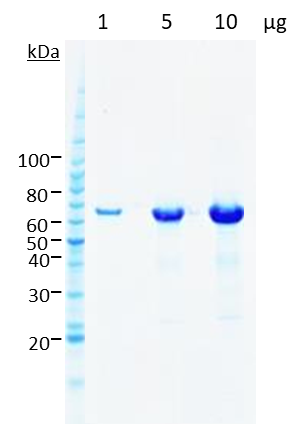
Five constructs were chosen in discussion with the client for scale-up and protein purification. In every case, between 1–10mg of homogenous protein at concentrations of 3–8mg/ml and a purity >95% (see Figures 4 and 5) were delivered to the client for preliminary structural studies.
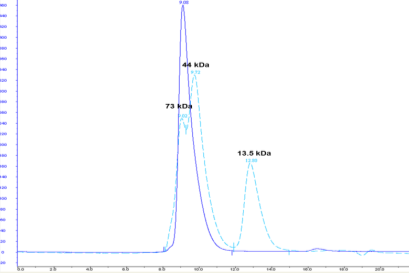
Conclusions
Our CDH technology enabled us to produce a large quantity of a protein that had previously been challenging, and impossible to make on significant scale. 70mg was purified in one batch and showed good activity in a biochemical assay. Our client was able to get a high-resolution X-ray crystal structure with one of the constructs, which enabled their SBDD programme for a target which had hitherto been refractory to crystallography.
Case Study 3: KMT2D: SET Domain via CDH
Histone-lysine N-methyltransferase 2D (KMT2D, also known as MLL4) is a member of a family of six histone H3K4-specific methyltransferases, the other members being KMT2A (or MLL1), KMT2B (or MLL2), KMT2C (or MLL3), KMT2F (or SET1A) and KMT2G (or SET1B). Each member of the family contains a catalytic SET domain, and other domains that mediate interactions with DNA and with other proteins. Mutations of KMT2D are associated with a number of diseases including congenital heart disease, various forms of cancer and Kabuki syndrome.
Challenge
Domainex collaborated with Dr Jon Wilson of the Francis Crick Institute to study the structure and activity of the SET domain of KMT2D. Previous attempts to produce KMT2D constructs in the Wilson lab had not yielded crystals despite extensive efforts.
Solution
Domainex used its proprietary CDH technology in order to identify constructs that contained both the SET and postSET domains of KMT2D. From a library of 157,000 clones, 18 unique, well-expressed and soluble constructs were identified that covered the required region (Figure 6).
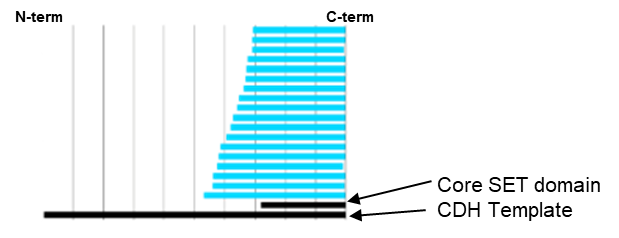
One of these CDH constructs was successfully used to obtain a high-resolution (2.2Å) crystal structure of the SET domain in complex with the cofactor S-adenosyl methionine (SAM) (Figure 7). The construct was shown to bind to SAM by differential scanning fluorimetry (DSF) analysis (Figure 8), but was found to be inactive in a biochemical assay. The crystal structure showed that the C-terminal tag occupied the substrate binding pocket, and this explained why the construct was catalytically inactive. We therefore expressed and purified the same construct but this time with an N-terminal tag and this protein was indeed shown to be catalytically active.
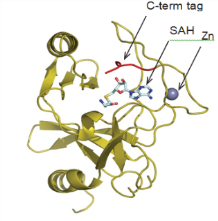
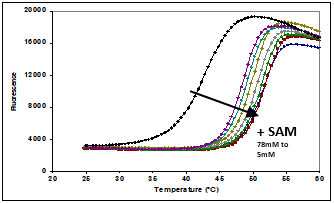
Conclusions
Our CDH technology allowed a high-resolution crystal structure of the SET domain of KMT2D to be obtained. This structure has revealed a mechanism for SET domain activation based on the structural differences between KMT2A and KMT2D.
This work resulted in a publication.3
Domainex Expertise
• Combinatorial Domain Hunting (CDH) • High Throughput Protein Expression • High Variant Protein Expression
• Protein Science • X-ray Crystallography • Structural Biology
References
- Fused thiophene derivatives as MEK inhibitors. Victoria E. Laing, Daniel C. Brookings, Rachel J. Carbery, Jose Gascon Simorte, Martin C. Hutchings, Barry J. Langham, Martin A. Lowe, Rodger A. Allen, Joanne R. Fetterman, James Turner, Christoph Meier, Jeff Kennedy and Mark Merriman. Bioorganic & Medicinal Chemistry Letters, 2012, 22, 1, 472-475
- Engineering human MEK-1 for structural studies: A case study of combinatorial domain hunting. Christoph Meier, Daniel C. Brookings, Thomas A. Ceska, Carl Doyle, Haiping Gong, David McMillan, Giles P. Saville, Adeel Mushtaq, David Knight, Stefanie Reich, Laurence H. Pearl, Keith A. Powell, Renos Savva, Rodger A. J Struct Biol. 2012, 177, (20), 329-34
- Evolving Catalytic Properties of the MLL Family SET Domain. Ying Zhang, Anshumali Mittal, James Reid, Stephanie Reich, Steven J. Gamblin and Jon R. Wilson. Structure 2015, 23, 1921–1933
Start your next project with Domainex
Contact one of our experts today