- About
-
Solutions
-
Services
- Biosciences
- Chemistry
- Integrated Drug Discovery
- Computer Aided Drug Design
- Hit Identification
- DMPK
- Target Classes and Modalities
- Therapeutic Areas
-
A-Z
- A
- B
- C
- D
- E
- F
- G
- H
- I
- K
- L
- M
- N
- O
- P
- R
- S
- T
- V
- X
-
Services
- Library
- News & Events
- Careers
Differential Scanning Fluorimetry (DSF) and nanoDSF Services
Characterising ligand induced changes in protein stability

Differential Scanning Fluorimetry (DSF) and nanoDSF (intrinsic DSF) are widely used, versatile biophysical techniques employed across diverse drug discovery applications, including fragment screening. The binding of even low molecular weight ligands can increase protein stability and can therefore be measured.
Differential Scanning Fluorimetry Services at Domainex
As part of our integrated drug discovery services, Domainex has invested in the Prometheus NT.Plex1 from NanoTemper2, a capillary-based instrument that measures protein folding in a label-free manner, using nanoDSF. We also have a Bio-Rad CFX OPUS3 for standard DSF measurements.
Differential Scanning Fluorimetry (DSF)
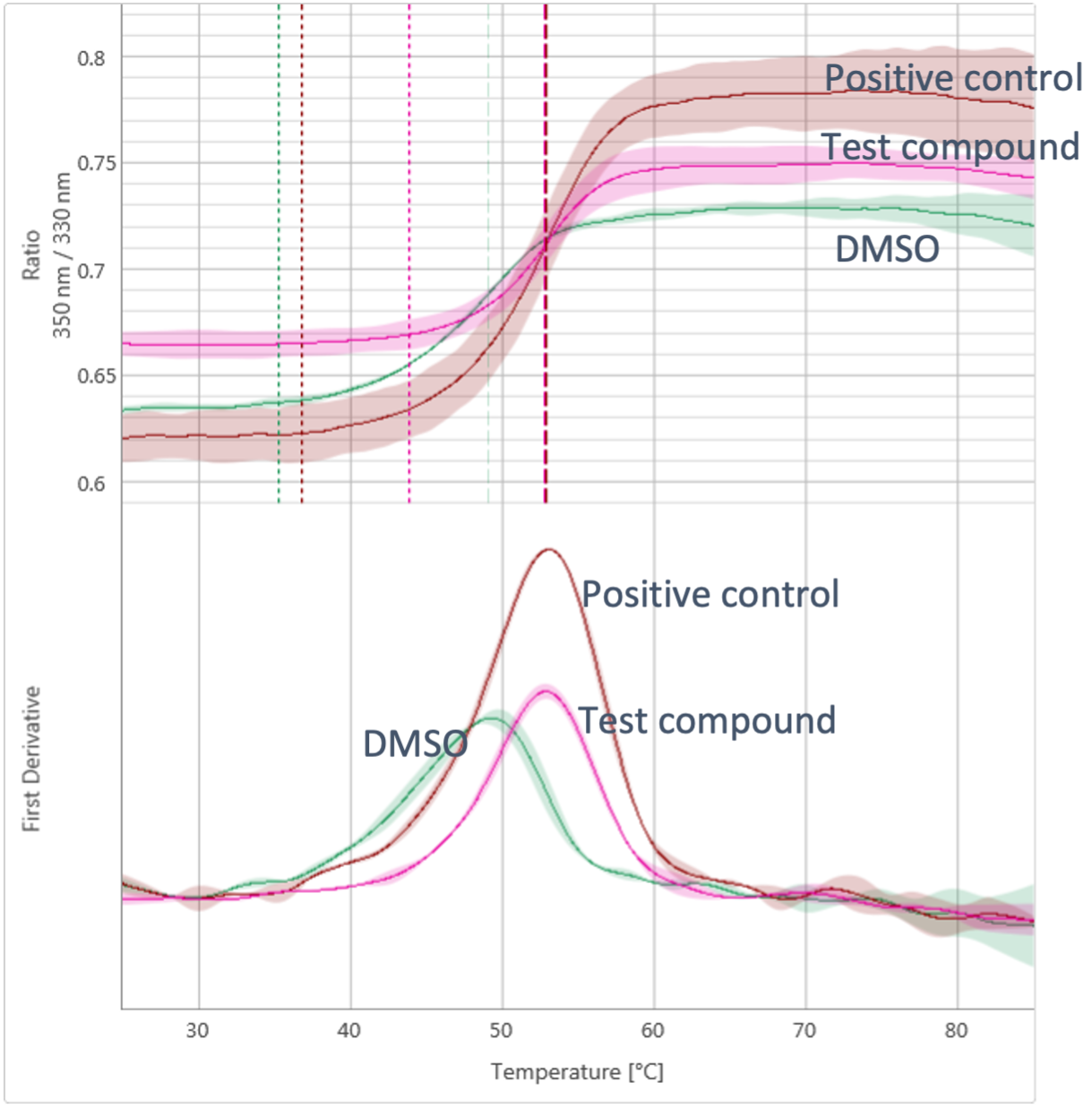
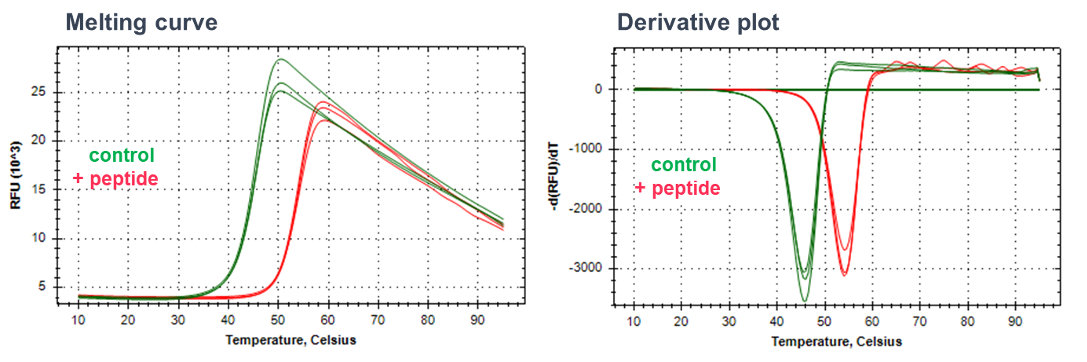
DSF is used to determine ligand binding by measuring ligand-mediated shifts in the melting temperature of the target protein. Purified protein is incubated with an environmental fluorescent dye (such as Sypro Orange), the fluorescence of which is quenched by water. The protein is exposed to a thermal gradient and as the protein unfolds, the hydrophobic residues are increasingly exposed, leading to an increase in dye binding and an increase in signal due to the exclusion of water. Fluorescence is measured continually, and compound binding is anticipated to stabilise the protein and shift the temperature at which the protein melts (measured as the temperature giving a half maximal effect’ Tm).
Advantages of DSF
- Choice of environmental dyes allows flexibility in measuring unfolding processes
- Very fast set-up (minutes)
- Medium throughput in 384 well plates
- Applicable to almost any type of globular protein (small or large), including enzymes, antibodies and antibody drug conjugates (ADCs)


nanoDSF
In the case of nanoDSF, rather than employing a dye, the intrinsic fluorescence of tryptophan and tyrosine residues in the protein is measured. As the protein unfolds the fluorescence intensity of these residues changes. nanoDSF has almost no restrictions in buffer composition, making it suitable for difficult to work with samples such as solubilised membrane proteins. As well as determining melting temperatures, the Prometheus NT.Plex has additional functionalities, including being able to measure aggregation as another method for analysing ligand binding. nanoDSF uses less protein than standard DSF, making it flexible and efficient, albeit with a lower throughput than traditional DSF.
Advantages of nanoDSF
- Label-free: measures intrinsic fluorescence of proteins
- Protein conservative
- Measures Tm, Tonset and Tagg in same experiment
- Wide range of buffers and viscous solutions can be used
- Applicable to almost any type of protein (small or large), including enzymes, antibodies, antibody drug conjugates (ADCs) and membrane proteins
Start your next project with Domainex
Contact one of our experts today