- About
-
Solutions
-
Services
- Biosciences
- Chemistry
- Integrated Drug Discovery
- Computer Aided Drug Design
- Hit Identification
- DMPK
- Target Classes and Modalities
- Therapeutic Areas
-
A-Z
- A
- B
- C
- D
- E
- F
- G
- H
- I
- K
- L
- M
- N
- O
- P
- R
- S
- T
- V
- X
-
Services
- Library
- News & Events
- Careers
Ternary Complex Formation Assays
The key to progressing your TCF project
At Domainex, our protein scientists have extensive experience of producing high-quality E3 ligase proteins (e.g. Cereblon and VHL) and offer a suite of assays to assess both binary and more importantly, ternary complex formation (TCF).
The use of small molecules to induce proximity between target proteins, thereby modulating their interactions and functions. This generates an exciting modality that is enabling the discovery of new drugs for challenging targets. One of the most notable applications of this approach is in the field of targeted protein degradation, particularly with PROTACs® (PROteolysis TArgeting Chimeras) and molecular glues.
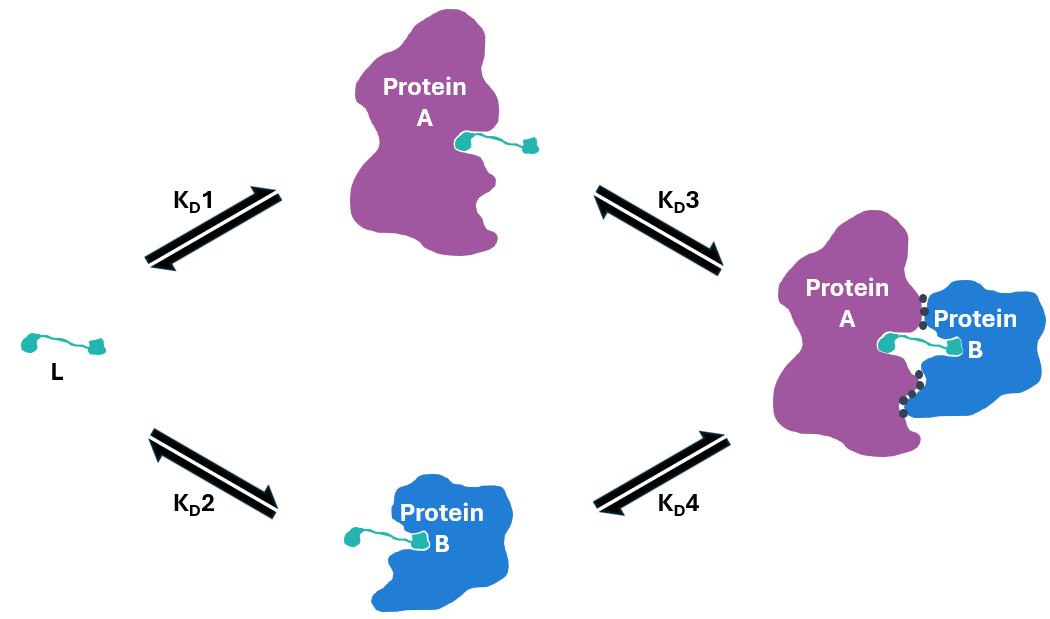
Figure 1: Ternary complex equilibria model1 for protein A, protein B and proximity-inducing ligand (L): Binary binding affinity of L and protein A (KD1); Binary binding affinity of L and protein B (KD2); Ternary complex binding affinity of protein A/L complex and protein B (KD3); Ternary complex binding affinity of protein B/L complex and protein A (KD4).
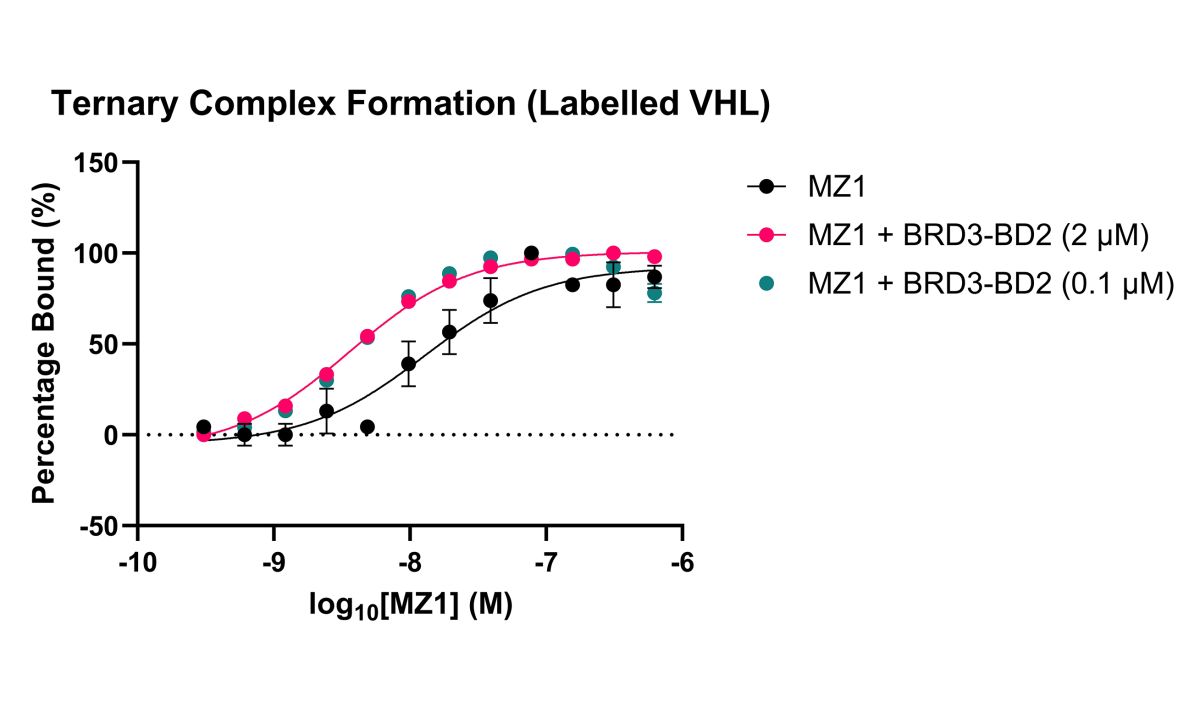
Figure 2: Spectral Shift assay measuring binary and ternary complex formation (positive cooperativity). Binary binding of MZ1 to fluorescently labelled VHL protein (black circles) and ternary complex formation (VHL, BRD3 and MZ1) measured at high (pink circles; 2 μM) and low (green circles 0.1 μM) BRD3 concentrations. Data is normalised as percentage bound based on the maximum binding signal for each experiment.
The small molecule induces or enhances ternary complex formation (TCF) between the two proteins of interest. Demonstration that TCF occurs is an important step in the discovery and characterisation of this class of therapies. Our in-house suite of assays allow us to quickly measure the affinity, co-operativity, kinetics and thermodynamics of TCF, all of which play crucial parts of drug design and optimisation.
The ternary complex equilibria model1 (Figure 1) describes the formation and dissociation of a ternary complex involving three components: Protein A, the small molecule ligand (L), and Protein B. This model provides a framework for understanding the dynamics and efficiency of interactions between these molecules. Measuring the binding affinity of each step in the formation of the ternary complex enables the determination of the cooperativity factor (a). For example, we can determine how protein B affects the interaction between L and protein A via the calculation of the cooperativity factor, a = KD1/KD4. Values >1 indicate positive cooperativity and enhancement of the complex, whereas values <1 indicates negative cooperativity and a reduction of the complex.
Domainex offers a variety of assay formats for the characterisation of TCF, and using our depth of experience developing such assays we can help you find the optimal assay for your specific programme.
Spectral Shift (SpS, NanoTemper)
- Microwell plate-based, solution assay format
- Low (μg) protein requirements per sample
- Medium throughput (10s – 100s of KD values can be determined each day )
- Compatible with our in-house Direct-to-Biology (D2B)
- Requires labelling of a single protein
- Determination of binary and ternary complex affinity
- Determination of co-operativity
- Suitable for medium throughput screening (e.g. fragment-based screening for molecular glue hit identification)
Find out more about our Spectral Shift offerings:
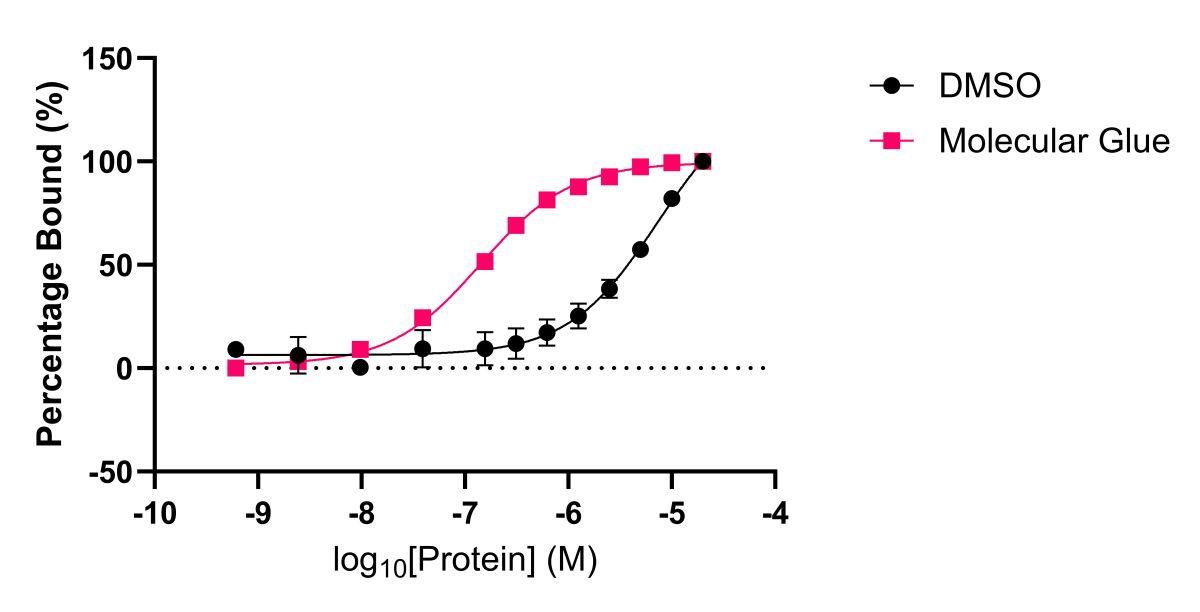
Figure 3a: Determination of the effect of a molecular glue on a protein-protein interaction via Spectral Shift
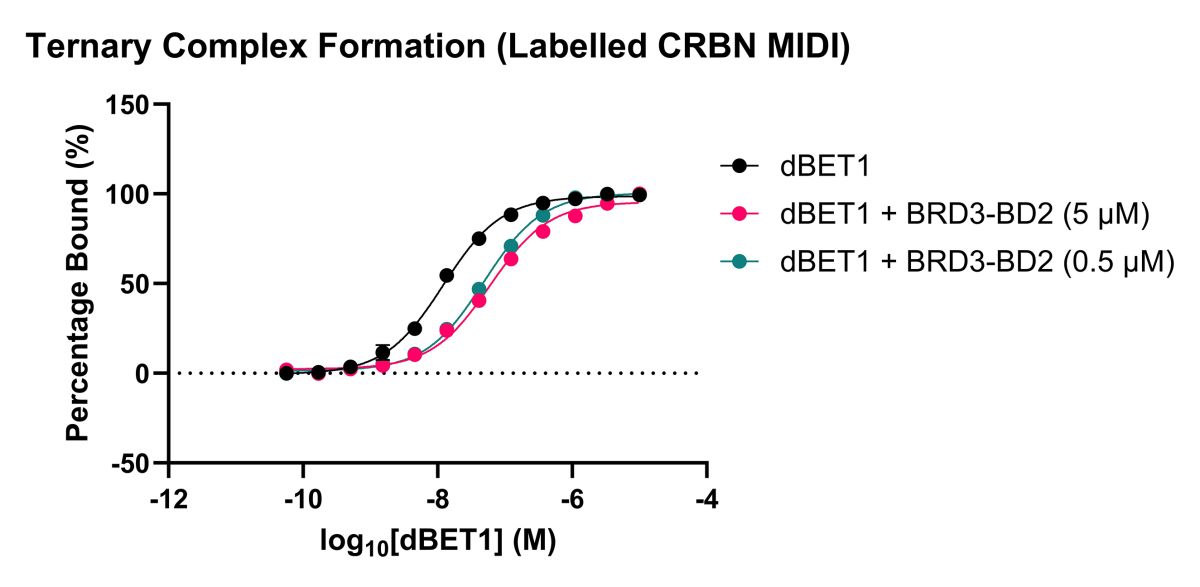
Figure 3b: Binary and ternary complex (in the presence of BRD3-BD2) binding curves for dBET1 to fluorescently labelled Cereblon midi protein via Spectral Shift
Grating Coupled Interferometry (GCI, Malvern Panalytical), a similar technique to Surface Plasmon Resonance (SPR, Cytiva)
- Chip-based formats with either E3 or Protein of Interest (POI) immobilised to the chip surface
- Low (μg) protein requirements for immobilised protein, higher quantities (μg to mg) for binding partner (analyte)
- Low throughput (1-10 compounds assessed per day)
- Compatible with most of our internal D2B formats
- Detailed characterisation including binding affinities and kinetics (kon & koff) and cooperativity and stability of TCF
Find out more about our GCI and SPR offerings:
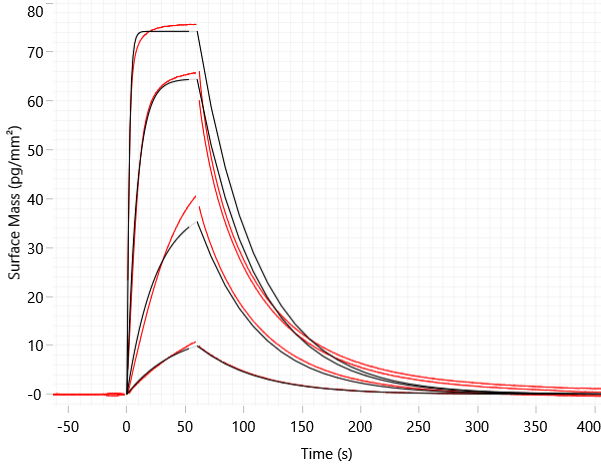
Figure 4: Assessment of ternary complex formation (BRD3 binding to immobilized VHL in the presence of MZ1) via GCI
Proximity Biochemical Assays: OMEGATM (Biosignal2), AlphaLISATM (Revvity), HTRFTM (Revvity)
- Microwell plate-based, solution assay format
- Low (ng) protein requirements per sample
- Requires labelling of both protein partners
- High throughput (10-1000s of KD values can be determined each day)
- Affinity determination of complex formation
- Suitable for high throughput screening for molecular glue hit identification
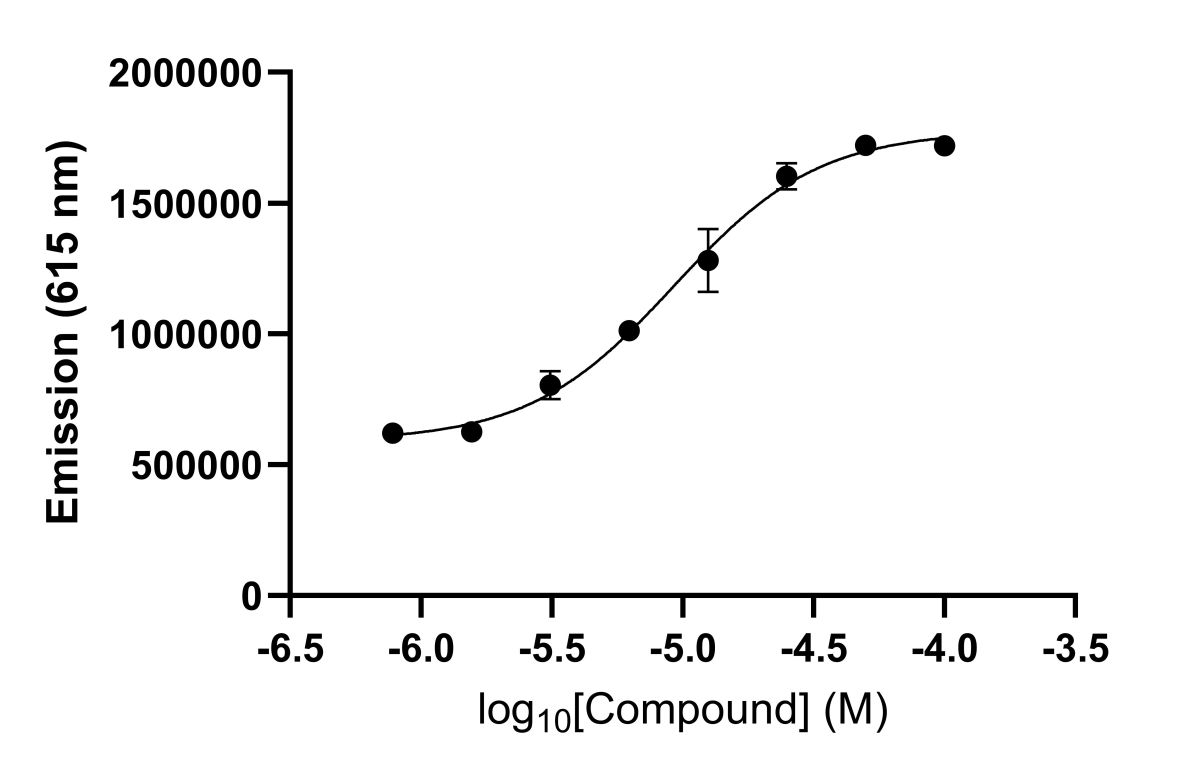
Figure 5: Determination of the EC50 of a molecular glue via AlphaLISA
Mass-Photometry (Refeyn)
- Orthogonal method for confirmation of TCF (no fluorescence, immobilisation or tags required)
- Solution-based, label-free format
- Low protein requirements (1-100 nM of protein)
- Low throughput (1-6 compounds assessed per day)
- Cannot determine binary binding affinity of small molecules
- Ternary complex affinity can be determined but data is limited by throughput and the protein concentration range
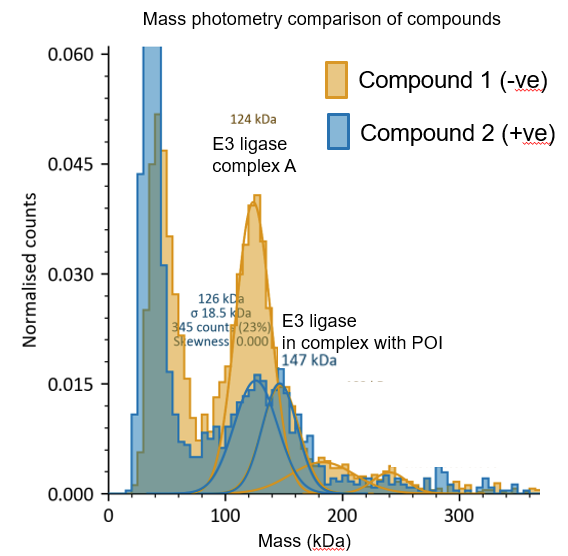
Figure 6: Assessment of ternary complex formation via mass photometry in the presence of a PROTAC® molecule
Isothermal Titration Calorimetry (ITC, Malvern Panalytical)
- Solution-based, label-free format
- High (mg) protein requirements
- Low throughput (1-2 compounds assessed per day)
- Characterisation of thermodynamics (ΔH, ΔS, and ΔG), binding affinities, stoichiometries, and cooperativity
Find out more about our ITC offerings:
PROTAC® is a registered trademark of Arvinas Operations, Inc., and is used under license.
Reference:
- Hughes, S. J. & Ciulli, A. Molecular recognition of ternary complexes: a new dimension in the structure-guided design of chemical degraders. Essays in Biochemistry 61, 505–516 (2017).
Start your next project with Domainex
Contact one of our experts today