By Dr Alicia Galván Álvarez (Team Leader, Chemistry)
PROTAC®s (PROteolysis TArgeting Chimeras) represent a therapeutic modality that targets specific proteins for degradation (by recruiting E3 ligases), leading to their degradation via the ubiquitin-proteasome system, offering the potential for treating diseases where protein dysregulation is a factor. The field of Targeted Protein Degradation (TPD) has drawn the attention of the scientific community in the last decade, with the number of publications rising exponentially. Despite recent advances in CNS degraders, design principles and assay cascades required for rapid CNS PROTAC® discovery are yet to be established. In general, identifying new chemical entities or hits as starting points within the PROTAC® field remains an empirical process where hundreds of compounds must be synthesised, purified and tested. In this field, “Direct-to-biology" (D2B) has recently emerged as a powerful new strategy that streamlines the synthesis and screening of compounds by directly evaluating crude reaction mixtures in biological assays, bypassing traditional purification steps.
Recent progress in this area from different research groups and companies has primarily been achieved by the use of pre-conjugated E3 ligase ligand-linker building blocks ready to be coupled to the protein of interest (POI) ligand, enabling the plate-based synthesis of compounds and subsequent testing in cellular degradation assays.
In this blog, we highlight recent work (published as a pre-print in ChemRxiv) from the Farnaby Group at the University of Dundee Centre for Targeted Protein Degradation (UK), in partnership with BioTechne (Tocris), in which they present the discovery of a CNS active GSK3 degrader using a new concept: “orthogonally reactive linkers” and a direct-to-biology approach1.
PROTAC® potency, kinetics and ADME profiles rely on the selection of ligands, linkers and type of chemistry used. In the publication, the authors hypothesise that having a method where all the components could be modified at once would present a major advantage during the hit finding phase of the program. To achieve this, they established the use of “orthogonally reactive linkers” as reagents that could be selectively coupled with high conversion at either end through two orthogonal reaction types in a stepwise fashion (Figure 1).
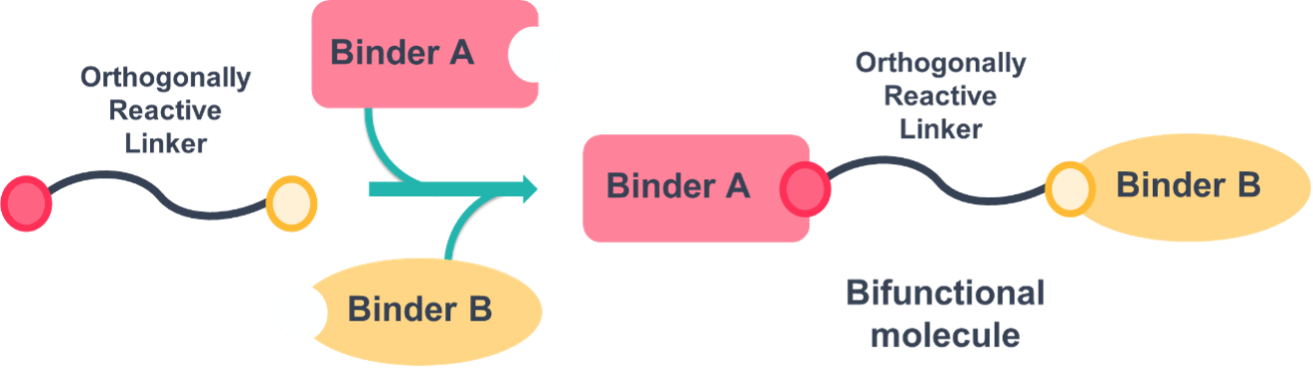
Figure 1: Orthogonally Reactive Linkers
As the focus of their library design was towards obtaining CNS active GSK3 degraders, they moved away from traditional amide couplings, with the aim of limiting the hydrogen bond donor (HBD) count and large increases in LogP. SN2 and CuAAC “click” reactions were chosen as they provide high conversion while limiting the increase in polar surface area and HBD count. To overcome some of the challenges encountered in previous studies with unreacted ligand reagents and other by-products, they introduced a basic centre in the linker to enable a plate-based SCX purification step. As the goal was to achieve CNS activity, they mainly introduced CRBN-recruiting ligands and retained only one VHL-recruiter.
The libraries were generated using a liquid handling system in a 96-well plate format performing an SN2 reaction, followed by the CuAAC, and plate-based SCX purification. Eluted compounds were then directly employed in cell-based high-throughput degradation screens using a GSK3β-HiBiT knock-in HEK293 cell line. The full workflow including synthesis of two 96-well plates (192 compounds) and cellular screening in 384-well plates was completed in 5 days (Figure 2).
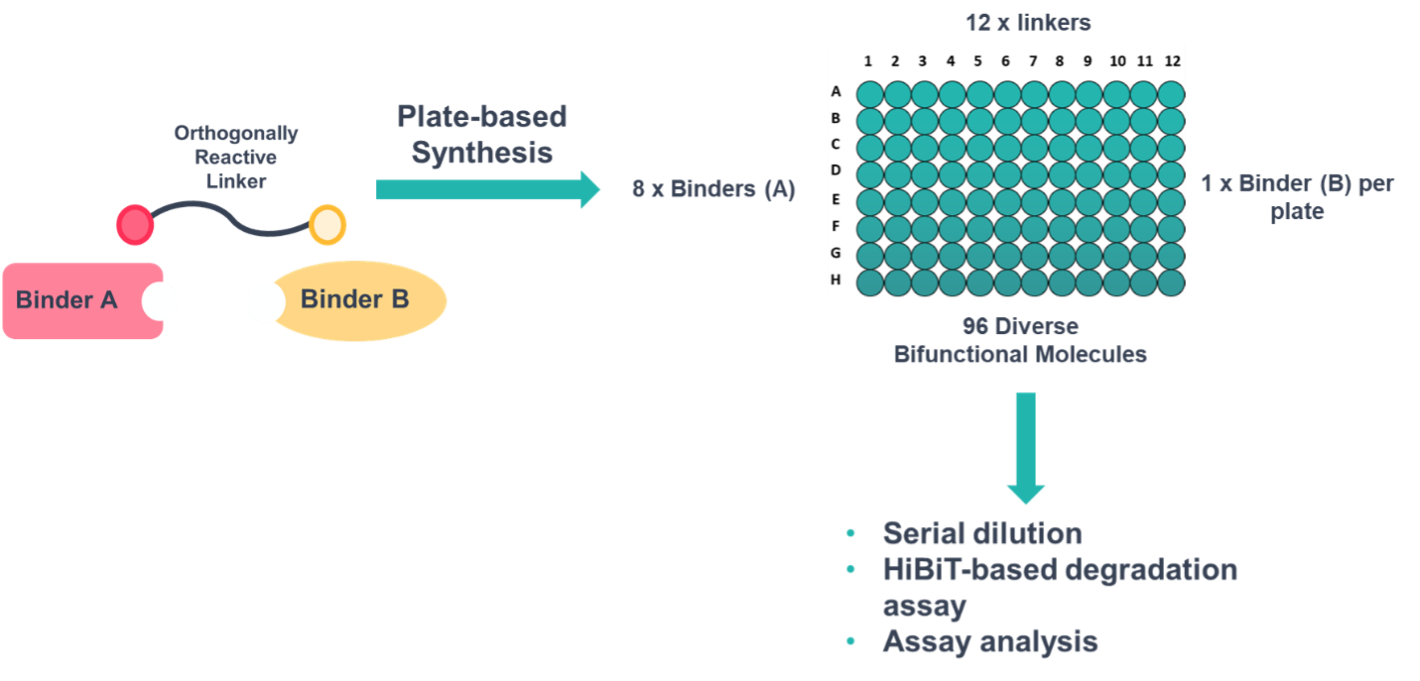
Figure 2: D2B screening platform using orthogonally reactive linkers
From a single experiment, a wealth of structure-activity relationship (SAR) data was generated. Six compounds with different linkers and E3 ligase ligands were selected, re-synthesised and purified, so the degradation profiles of crude vs purified compounds could be compared. This confirmed that they retained their high levels of degradation activity. The power of this new approach is demonstrated, as from just one orthogonally reactive linker D2B screen they were able to identify degraders from multiple series. From the six resynthesised compounds, they identified two molecules of particular interest, KH1 (24) and KH2 (26), which had DC50 values in the picomolar to single digit nanomolar range and high selectivity for degradation of both GSK3 paralogs with no other significant degraded proteins identified.
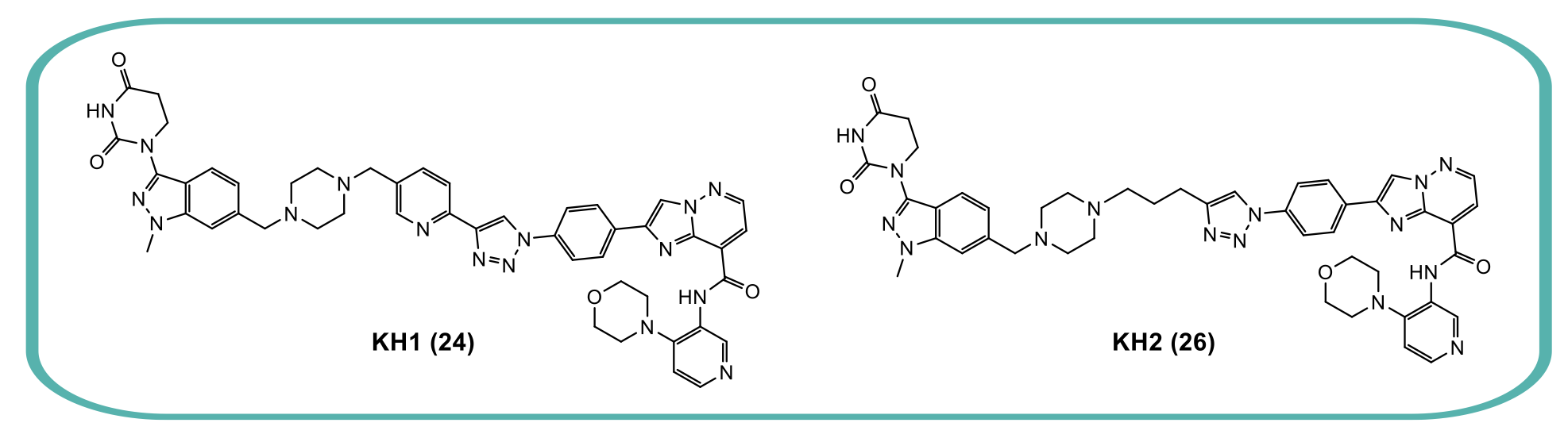
Figure 3: The structures of KH1 (24) and KH2 (26).
In vivo studies showed that following a single 5mg/kg i.v. dose, KH1 was able to almost completely remove GSK3β from the liver with partial degradation also observed in mouse brain. This was achieved by the combination of moderate brain penetration and potent and rapid degradation kinetics.
The work discussed here has tackled different challenges at the same time, developing a novel orthogonal and high-throughput platform to identify new degraders in a D2B fashion while identifying a CNS-active GSK3 degrader. This opens the door to exploring alternative ways of investigating GSK3 paralogs in neurodegenerative diseases.
At Domainex we have extensive experience in D2B and we have generated a PROTAC® toolbox designed to be combined with our D2B platform for the rapid, plate-based synthesis and evaluation of PROTAC®s. Whether it’s to enable your Targeted Protein Degrader (TPD) project or supercharge your hit-to-lead (H2L) or lead optimisation (LO) programmes, Direct-to-Biology will open up a galaxy of opportunity for you! So, partner with Domainex and revolutionise your drug development process today.
If you’d like to know more about D2B and its potential, you can watch our on-demand Webinar and see our other related blogs: An introductory guide to Direct-to-Biology and Direct-to-biology (D2B) beyond amide couplings. Please get in touch if you would like to find out more, we’d love to hear from you!
PROTAC® is a registered trademark of Arvinas Operations, Inc., and is used under license
References:
- Discovery of a CNS active GSK3 degrader using orthogonally reactive linker screening. Andreas Holmqvist , Nur Mehpare Kocaturk, Christina Duncan, Jennifer Riley, Steve Baginski, Graham Marsh, Joel Cresser-Brown, Hannah Maple, Kristiina Juvonen, Nicola Morrice, Calum Sutherland, Kevin D. Read and William Farnaby*. ChemRxiv. 2025; doi:10.26434/chemrxiv-2025-nq14w