- About
-
Solutions
-
Services
- Biosciences
- Chemistry
- Integrated Drug Discovery
- Computer Aided Drug Design
- Hit Identification
- DMPK Services
- Target Classes and Modalities
- Therapeutic Areas
-
A-Z
- A
- B
- C
- D
- E
- F
- G
- H
- I
- K
- L
- M
- N
- O
- P
- R
- S
- T
- V
- X
-
Services
- Library
- News & Events
- Careers
RuvBL1/2: Cryo-EM analysis of a hetero-hexameric AAA+ ATPase
Generation of a cryo-EM structure to support structure-based drug design
Challenge
The phosphatidylinositol 3-kinase-related kinases (PIKK) family of protein kinases (members include ATR, ATM, SMG1 and mTOR) are key targets in the hunt for novel cancer therapeutics. RuvB-like proteins 1 and 2 (RuvBL1/2) are two highly conserved AAA+ ATPases that assemble to form a hetero-hexameric complex. RuvBL1/2 (which forms part of the R2TP complex) has been observed to play a key role in assembly and maturation of large molecular PIKK family complexes such as mTOR1. Studies of RuvBL1/2 support a feedback mechanism between regulation of nucleotide access and interaction with other proteins to alter its hetero-hexameric ring conformation1. The RuvBL1/2 nucleotide-binding site is therefore of interest for drug design, and targeting RuvBL1/2 upstream of the protein maturation cascade could be a beneficial approach for small molecule drug development.
A high-resolution structure of the nucleotide-binding site was therefore required. The inherent flexibility in the central region of RuvBL1/2 makes this a challenging target for structural biology. Higher resolution (2.5A) has been achieved for crystallographic structures compared to cryo-EM structures, however protein truncation was required2. The advantage, therefore, of a Cryo-EM resolved structure is that the full complex is present, allowing allosteric effects of a compound, or binding partner, to be seen within the ATPase domain with a conformational change to restrict nucleotide access. Therefore Domainex selected this approach as the preferred option to support structure-based drug discovery (SSBD).
Solution
First, cryo-EM grid freezing conditions were optimised. Cryo-EM grids with consistent protein distribution over a sufficient area were progressed for data collection on a 300 KeV Titan Krios microscope with K3 detector. A total of 4,800 micrographs were collected for data processing.
3D processing of the full complex indicated a higher local resolution towards the hexametric rings of the head domain . In 2D class averages, alpha helices were observed within the ATPase domain (figure 1), confirming the presence of high-resolution information within the dataset. The team targeted the ATPase domain of the protein using focused processing, greatly improving the local resolution around the nucleotide-binding site. In the final maps, side chains were visible allowing reconstruction of the nucleotide-binding site and docking of adenosine diphosphate (ADP).
The RuvBL1/2 high-resolution map allows for visualisation of the ligand binding mode, enabling SBDD.
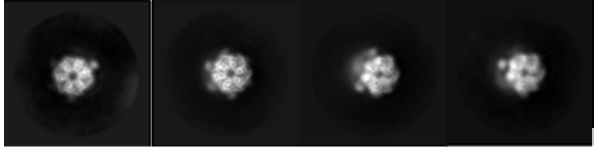
Figure 1: Selection of 2D class averages of RuvBL1/2 produced in RELION. Multiple views are seen of the complex with helices present in the ATPase domain from top-down views indicating high resolution information available.
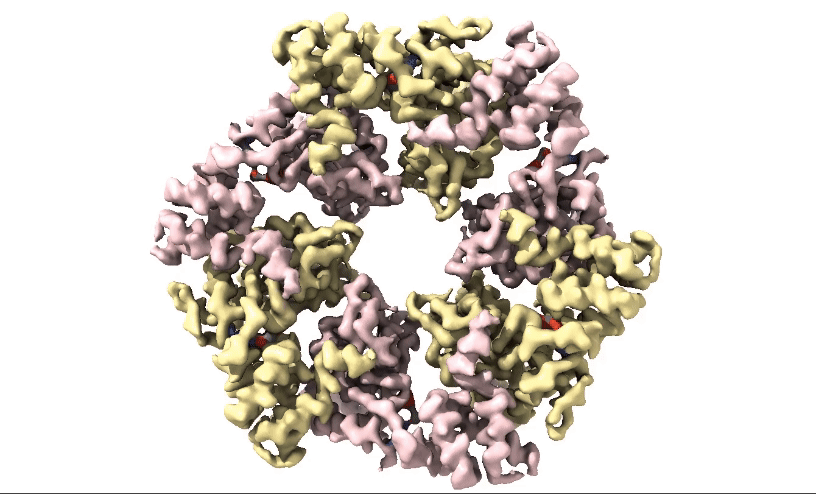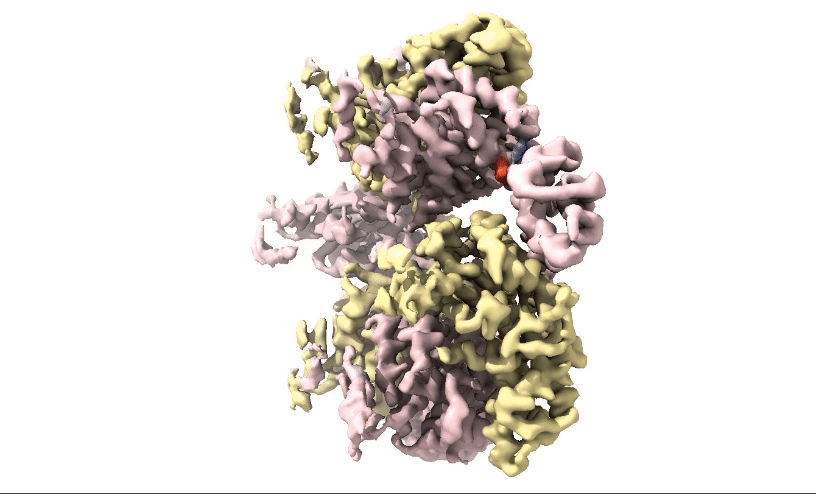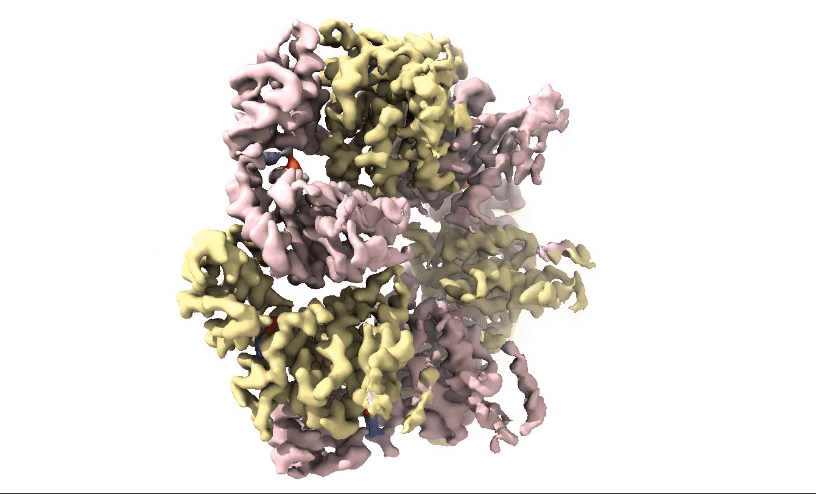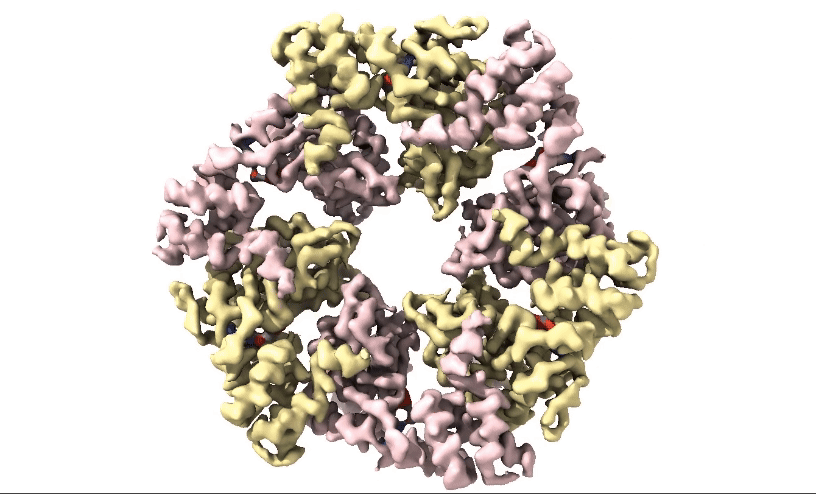
Figure 2: 3D 3.3Å Map of RuvBL1/2 visualised in ChimeraX. Protein is colour coded by RuvB monomer with RUVBL1 monomers in yellow and RUVBL2 monomers in pink. ADP is seen in the active site coloured in red.
Conclusion
Structural biologists at Domainex generated a 3.3 Å cryo-EM map of RuvBL1/2 (figure 2) using a RELION pipeline. The structure of the RuvBL1/2 nucleotide-binding site is clearly defined, with individual interactions between amino acid side chains and bound nucleotide observed, thereby opening the way for future SBDD efforts. This method can be applied to other targets to focus on the site of compound binding while maintaining the native structure of the protein.
Acknowledgements and References
RuvBL1/2 protein for the structural studies was supplied by Dr. Mohinder Pal (University of Kent) and Professor Laurence Pearl (University of Sussex).
Domainex utilised the Cryo-EM facility at the Cambridge University, Department of Biochemistry.
1: Muñoz-Hernández H, Pal M, Rodríguez CF, Fernandez-Leiro R, Prodromou C, Pearl LH, Llorca O. Structural mechanism for regulation of the AAA-ATPases RUVBL1-RUVBL2 in the R2TP co-chaperone revealed by cryo-EM. Sci Adv. 2019 May doi: 10.1126/sciadv.aaw1616.
PMID: 31049401; PMCID: PMC6494491.
2: Dapeng Ju et al., Chemical perturbations reveal that RUVBL2 regulates the circadian phase in mammals. Sci. Transl. Med.12, eaba0769(2020).DOI:10.1126/scitranslmed.aba0769
Start your next project with Domainex
Contact one of our experts today