At Domainex, we are continuously expanding our computational chemistry toolkit to better guide molecular design and selection. This quarter, we spotlight three new predictive approaches that directly address some of the most persistent challenges in drug discovery; solubility, permeability, and efflux: SMID (Smallest Maximum Intramolecular Distance), 3D Polar Surface Area (3DPSA), and pKBHX (Hydrogen-Bond Basicity). These complementary descriptors provide new ways to understand and optimise molecular behaviour, particularly for compounds that push beyond classical 'rule of 5' guidelines.
SMID – Chameleonic Behaviour and Intramolecular Shielding
The Smallest Maximum Intramolecular Distance (SMID) quantifies molecular compactness by measuring the maximum separation between heavy atoms. Molecules with low SMID values are able to adopt compact conformations that cloak hydrogen-bond donors and acceptors, revealing chameleonic behaviour that enhances permeability in nonpolar environments without permanently compromising solubility. SMID offers a valuable structural perspective on the critical balance between solubility and permeability, particularly relevant in modern lead optimisation programmes.
Recent work by Baylon et al.1 at Bristol-Myers Squibb introduced the Balanced Permeability Index (BPI), a composite metric that combines size, polarity, and lipophilicity, and improves oral bioavailability predictions for ligand-directed degraders (LDDs) such as PROTACs® (PROTAC® is a registered trademark of Arvinas Operations, Inc., and is used under license). In the study, SMID was incorporated into an augmented index, BPI_LDD, which accounts for cross-sectional molecular compactness. The addition of SMID significantly enhanced the ability to differentiate orally bioavailable degraders; BPI_LDD outperformed traditional descriptors (e.g., polarity, lipophilicity, size) and even some in vitro permeability measurements in predicting oral bioavailability of LDDs.
At Domainex, we have implemented SMID to complement our high-throughput ChromLogD platform, leveraging it as a pivotal parameter to optimise both permeability and solubility in PROTAC discovery projects. Together, ChromLogD and SMID serve as complementary tools; ChromLogD delivers rapid lipophilicity and polarity profiling, while SMID provides structural insight into molecular compactness; offering a powerful strategy in the design of orally bioavailable PROTACs.
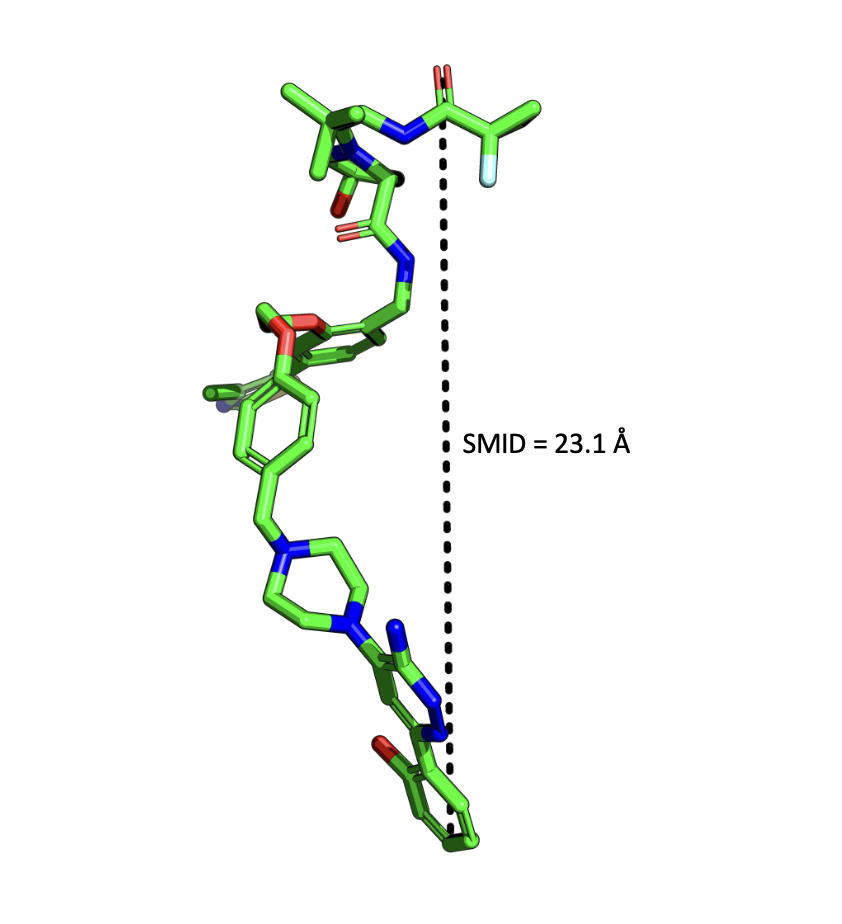
Figure 1. Example output from Domainex SMID calculation
3DPSA – Predicting Permeability Beyond the Rule of 5
Traditional PSA is a 2D descriptor that often underestimates how conformational dynamics influence polarity. Our 3DPSA workflow overcomes this limitation by averaging polar surface exposure across multiple conformers, providing a more realistic representation of the behaviour of flexible and larger molecules.
Why it matters: 3DPSA is a particularly powerful predictor of permeability for ‘beyond Rule of 5’ (bRo5) compounds such as macrocycles and PROTACs. By accounting for conformational flexibility, 3DPSA helps identify scaffolds that can fold to minimise polar exposure in membranes, while still maintaining solubility in aqueous media.
At Domainex, we were inspired by the pioneering work of Price et al.2 (Abbvie), who demonstrated the value of considering three-dimensional polarity in drug discovery. Building on this, we developed our own physics-based model for predicting 3DPSA/EPSA, drawing on the methodology described by Sebastiano et al.3
We have extensively benchmarked our approach against the challenging dataset published by Dr Henrik Möbitz4 (Novartis), which focuses on complex, highly flexible bRo5 molecules. Our method demonstrates very strong rank correlation in predicting permeability-relevant polarity across this dataset, highlighting its robustness and translational value for real-world drug discovery projects.
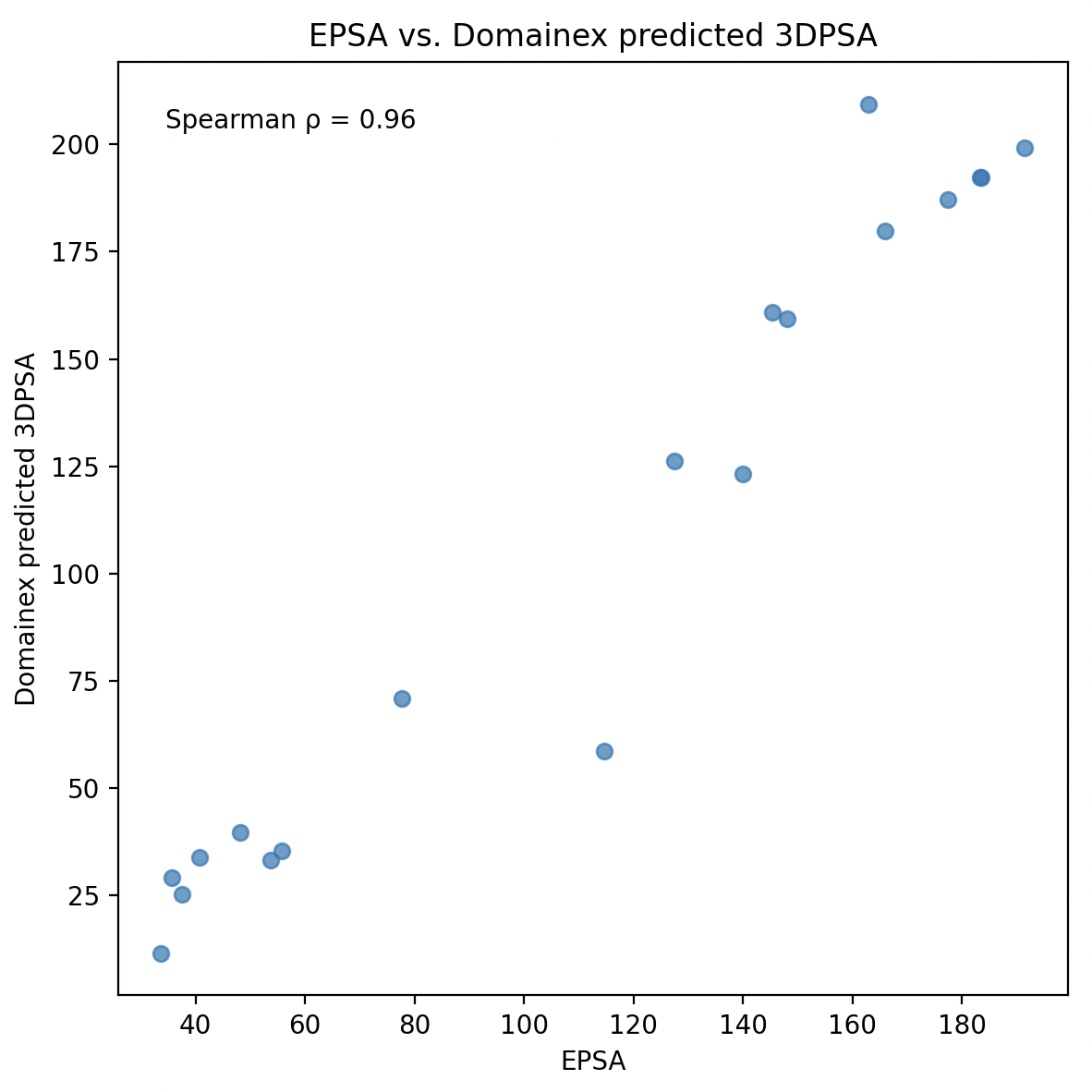
Figure 2. Comparison of EPSA vs. Domainex predicted 3DPSA for challenging dataset reported by Dr Henrik Möbitz
pKBHX – Optimising Efflux and ADME Balance
The hydrogen-bond basicity constant (pKBHX) quantifies a molecule’s ability to act as a hydrogen-bond acceptor. Compounds with elevated hydrogen-bond basicity may be susceptible to recognition by efflux transporters, reducing effective exposure. By optimising pKBHX, we can strike a balance in polarity, preserving permeability and solubility while mitigating efflux liability. Furthermore, this metric aids in fine-tuning selectivity and managing off-target interactions by tempering polar interactions.
pKBHX was first introduced by Peter W. Kenny et al.5 in their 2016 work, which established that minimised molecular electrostatic potential (V_min) is a reliable predictor of hydrogen-bond basicity and presented predictive models for a range of acceptor types.
Domainex was inspired by a recent blog post from Rowan Scientific,6 which described an efficient, physics-based artificial intelligence/machine learning (AI/ML) workflow for predicting site-specific hydrogen-bond acceptor strength. Building on that concept, we have developed our own proprietary physics-based AI/ML method to approximate pKBHX, which has been extensively benchmarked against literature data with highly promising results.
Table 1. Comparison of Rowan pKBHX values to Domainex’s proprietary pKBHX method.
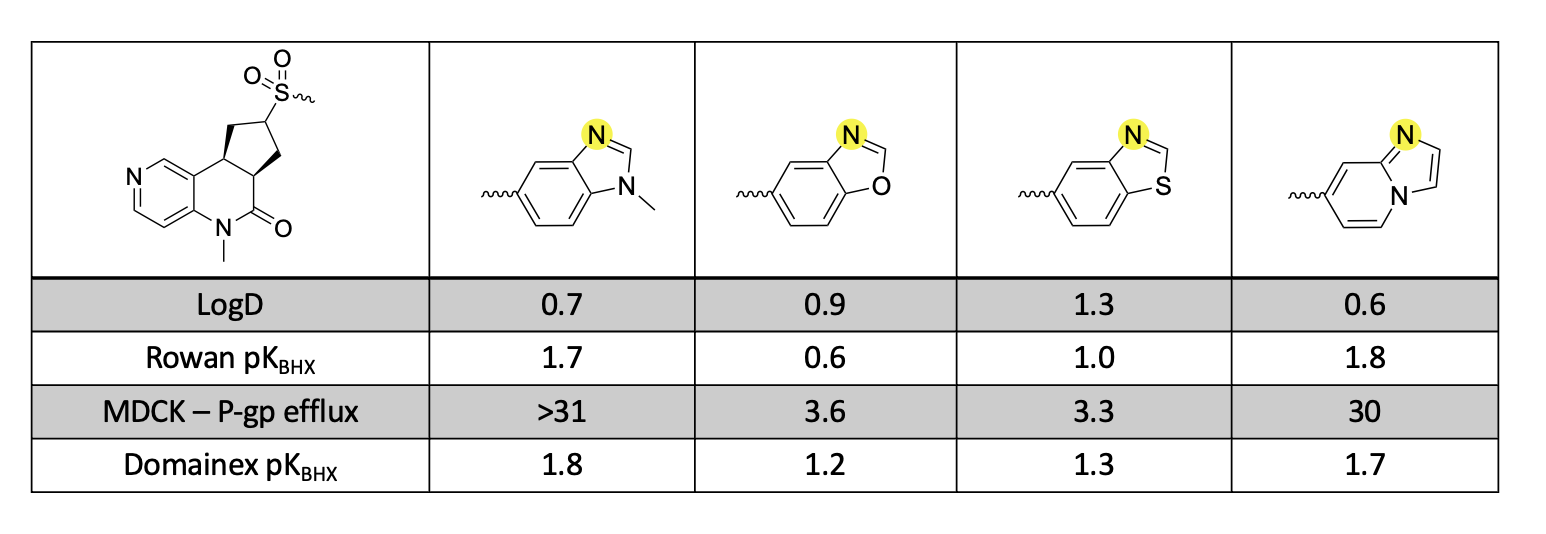
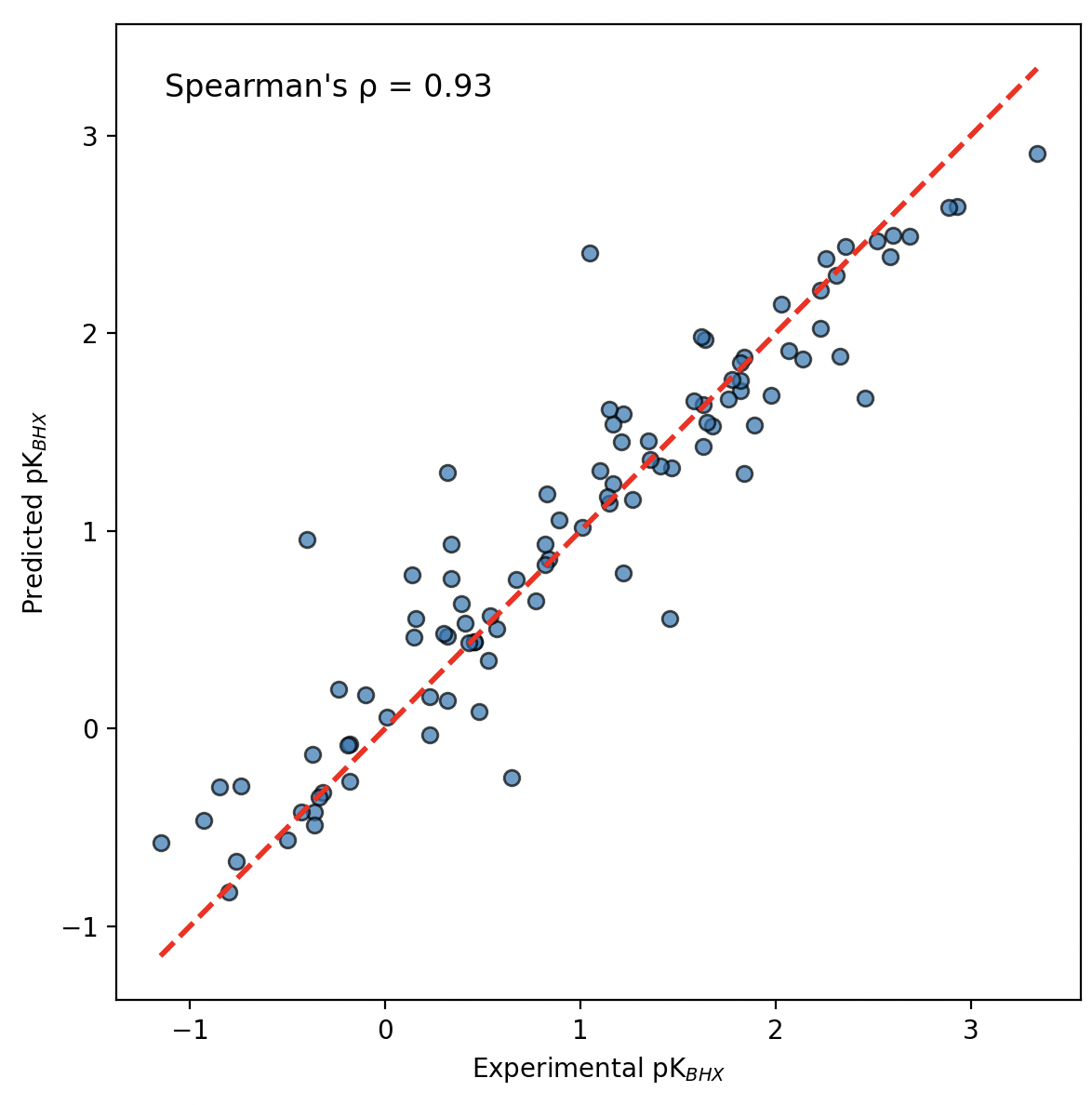
Figure 3. Comparison of 100 experimental pKBHX values to the predicted pKBHX from Domainex’s proprietary pKBHX method.
This enables Domainex to integrate pKBHX into rational optimisation of efflux and ADME balance in drug-discovery projects.
Integrated Impact
Together, these new tools provide a multi-dimensional view of molecular properties: SMID guides design of compact, chameleonic molecules; 3DPSA improves permeability predictions, especially for large and flexible scaffolds; pKBHX enables targeted optimisation of efflux and ADME balance. By integrating these descriptors, Domainex enhances its ability to design compounds with the right balance of solubility, permeability, and drug-like behavior, supporting faster, smarter progression from hits to clinical candidates.
Outlook
We’re already putting these predictive models to work on live discovery projects. In our upcoming blog posts, we’ll showcase real case studies where Domainex is using cutting-edge AI/ML and physics-based methods to accelerate drug design. Want to learn more? Get in touch with us today: Contact Domainex.
References:
- New Multiparameter Index Is a Strong Predictor of Oral Bioavailability for Heterobifunctional Degraders. Javier L. Baylon, Matthew J. Chalkley, Tony Siu, Wilson Shou, Yongnian Sun, Xianmei Cai, Anthony Paiva, Shivani Patel, Tatyana Zvyaga, and Dahlia R. Weiss. ACS Medicinal Chemistry Letters 2025 16 (6), 1108-1113.
- Explainable Machine Learning for ETR and Drug Chameleonicity. Edward Price, Matthieu Dagommer, Mattson Thieme, Richard Hong, J. Cory Kalvass, Stella Doktor, Alexey Rivkin, Yue-Ting Wang, Philip Cox, Abhishek Pandey, and David DeGoey. Journal of Medicinal Chemistry 2025 68 (15), 15636-15648.
- Impact of Dynamically Exposed Polarity on Permeability and Solubility of Chameleonic Drugs Beyond the Rule of 5. Matteo Rossi Sebastiano, Bradley C. Doak, Maria Backlund, Vasanthanathan Poongavanam, Björn Over, Giuseppe Ermondi, Giulia Caron, Pär Matsson, and Jan Kihlberg. Journal of Medicinal Chemistry 2018 61 (9), 4189-4202.
- H. Möbitz, ChemMedChem., 2024, 19, e202300395.
- Hydrogen Bond Basicity Prediction for Medicinal Chemistry Design. Peter W. Kenny, Carlos A. Montanari, Igor M. Prokopczyk, Jean F. R. Ribeiro, and Geraldo Rodrigues Sartori. Journal of Medicinal Chemistry 2016 59 (9), 4278-4288.
- https://www.rowansci.com/publications/hydrogen-bond-acceptor-strength-prediction