- About
-
Solutions
-
Services
- Biosciences
- Chemistry
- Integrated Drug Discovery
- Computer Aided Drug Design
- Hit Identification
- Target Classes and Modalities
- Therapeutic Areas
-
A-Z
- A
- B
- C
- D
- E
- F
- G
- H
- I
- K
- L
- M
- N
- O
- P
- R
- S
- T
- V
- X
-
Services
- Library
- News & Events
- Careers
Mass Photometry
Label-free protein oligomer detection
Principle of Operation
Mass photometry quantifies the interference between light reflected from a glass surface and light scattered by individual biomolecules transiently interacting with that surface.
The resulting contrast signal scales directly with molecular mass (Young et al., 2018), enabling accurate mass determination without labels or immobilization.
Applications
Protein State & Stability
- Oligomeric state determination (monomer → higher-order assemblies)
- Buffer and pH dependent stability profiling
- Assessment of stabilizing/destabilizing compound effects
Complex Formation
- Label-free detection of binary and ternary complexes, including those induced by bivalent compounds and molecular glues
- Quantification of compound-driven complex formation
- pH dependent complex formation analysis
Stability-monitoring of biomolecular complexes
Antibody & Nucleic Acid Systems
- Fragmentation and aggregation analysis
- Antibody–antigen binding assessment
- Protein–nucleic acid complex characterization
Key Advantages
- Native, in solution measurements
- Label-free detection
- Sensitive to sample heterogeneity and purity
- Minimal sample consumption (100 pM–100 nM)
- Broad mass range: 30 kDa–5 MDa
- Time-resolved measurements for dynamic processes
- Suitable for oligomerization and complex formation studies
- Compatible with membrane proteins
- Rapid data acquisition and turnaround
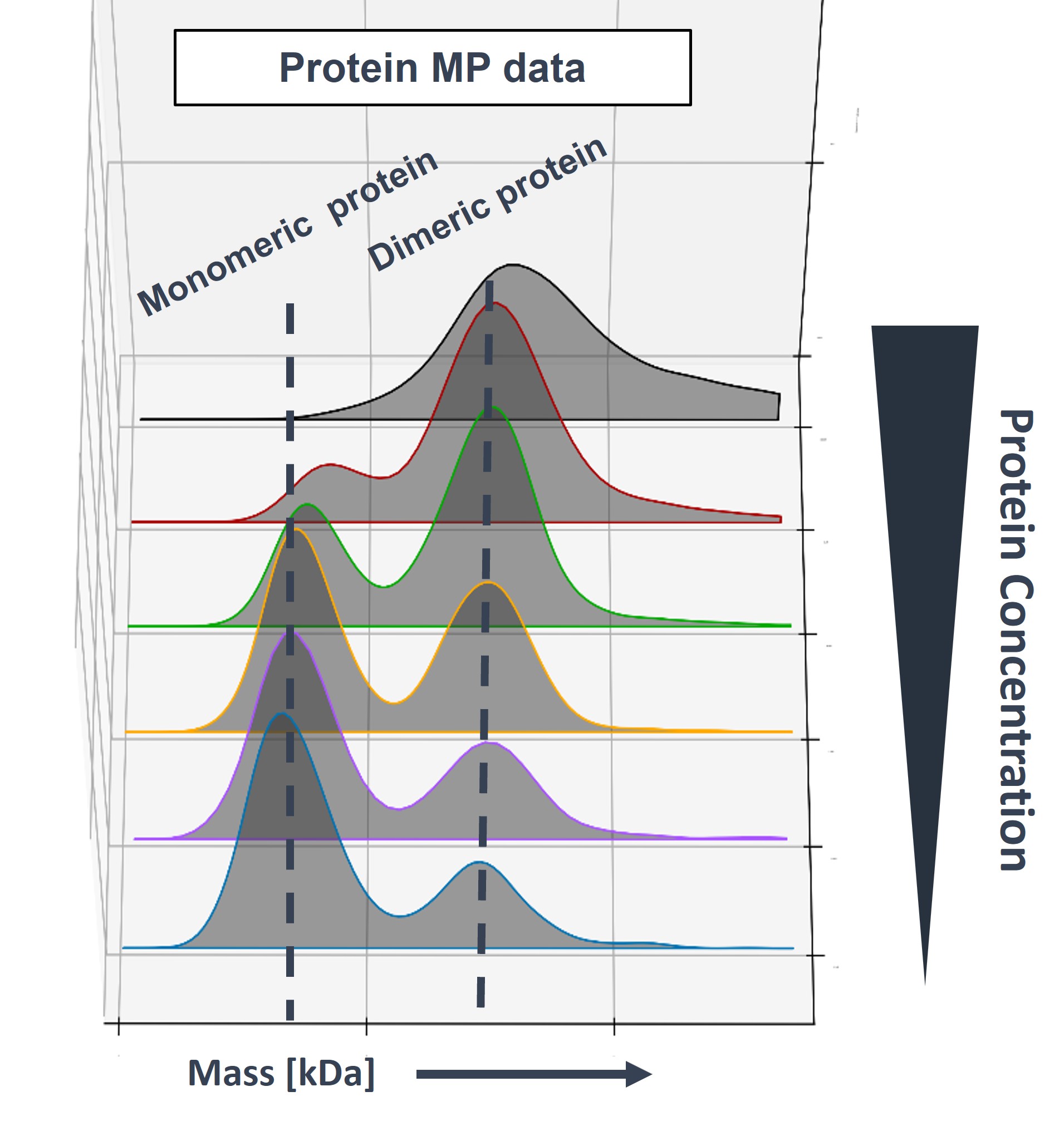
Example output showing protein dimer formation as concentration increases
Start your next project with Domainex
Contact one of our experts today