- About
-
Solutions
-
Services
- Biosciences
- Chemistry
- Integrated Drug Discovery
- Computer Aided Drug Design
- Hit Identification
- Target Classes and Modalities
- Therapeutic Areas
-
A-Z
- A
- B
- C
- D
- E
- F
- G
- H
- I
- K
- L
- M
- N
- O
- P
- R
- S
- T
- V
- X
-
Services
- Library
- News & Events
- Careers
G9a: A fragment-based drug design (FBDD) programme to identify lysine methyltransferase inhibitors for the treatment of cancer
Domainex successfully used MicroScale Thermophoresis to screen a sub-set of our fragment library against the lysine methyltransferase, G9a, with a hit rate of 5.3%.
Challenge
Lysine methyltransferases (KMTs) are involved in epigenetic gene regulation. Histones are proteins around which the DNA is wrapped in chromatin, and covalent modification of the histone amino-acid side chains generates an epigenetic ‘code’ that determines whether the associated gene is expressed or repressed. KMTs catalyse the transfer of methyl groups from S-adenosyl methionine (SAM) to lysine residues on histone proteins (Figure 1). G9a mono- or di-methylates Histone 3 at Lysine residue 9 (H3K9) and this represses gene expression. Literature supports the role of G9a in mechanisms of carcinogenesis, making it an attractive oncology target.1-5

Solution
Domainex has an extensive fragment library which contains >1000 fragments and has been carefully designed to maximise the chances of finding suitable chemical starting points (for more details on our library see our FragmentBuilder page). As a proof of concept study, a randomly selected 320 compound sub-set of this fragment library was screened at 1 mM against a G9a-SAM complex using MicroScale Thermophoresis (MST, see our Biophysics page for more details about this technique and our other biophysics services). By using a saturating concentration of SAM, we ensured that the co-factor binding pocket was not available for fragment binding, and so we specifically identified substrate-competitive hits. Fragments were declared as hits if a significant shift in the response compared to the reference was observed (Figure 2).
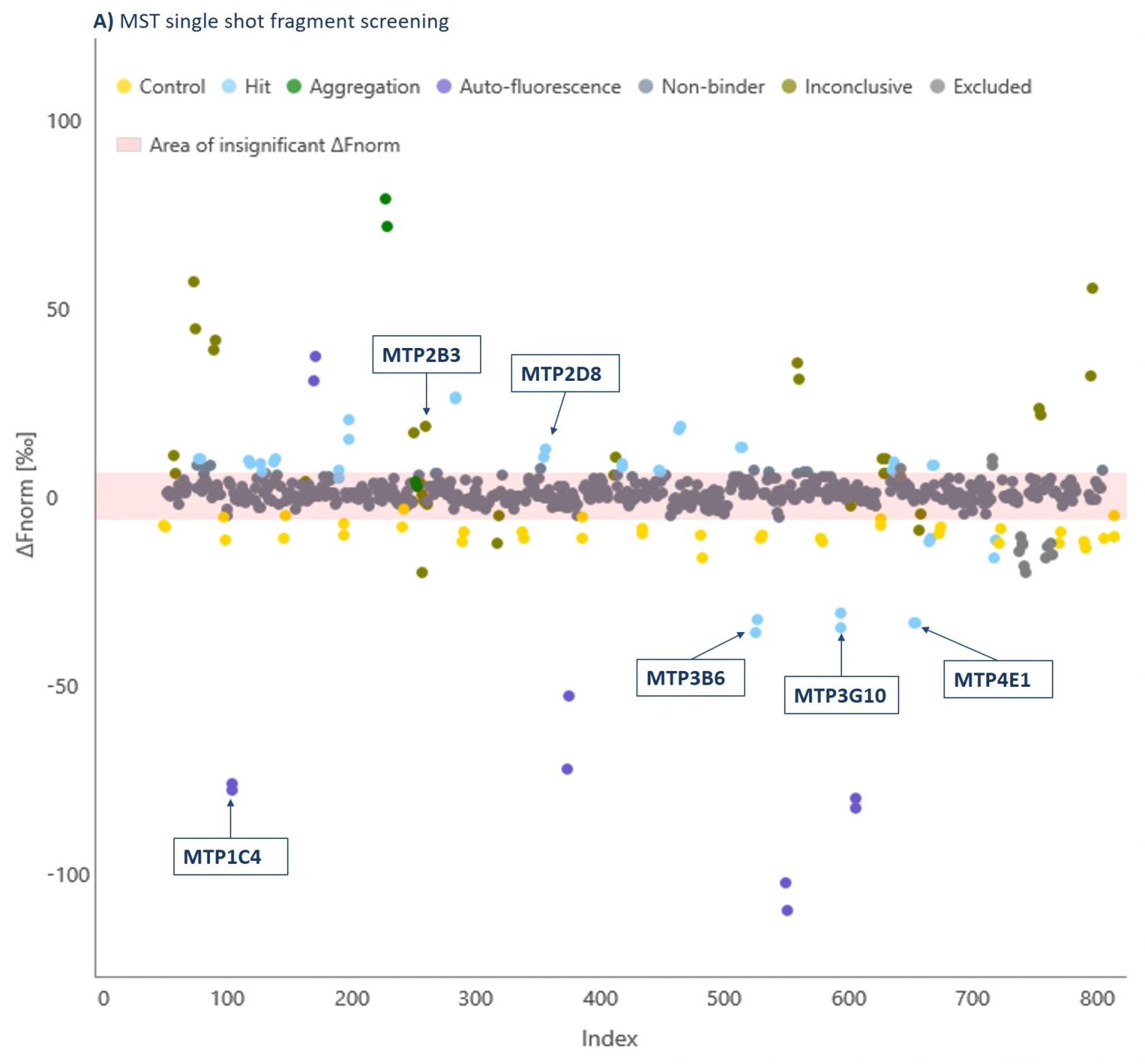
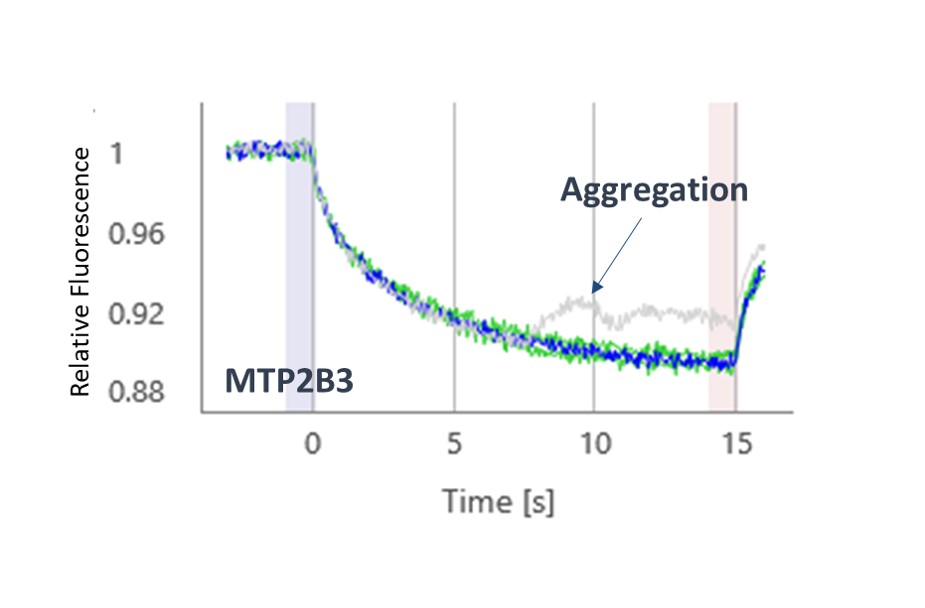
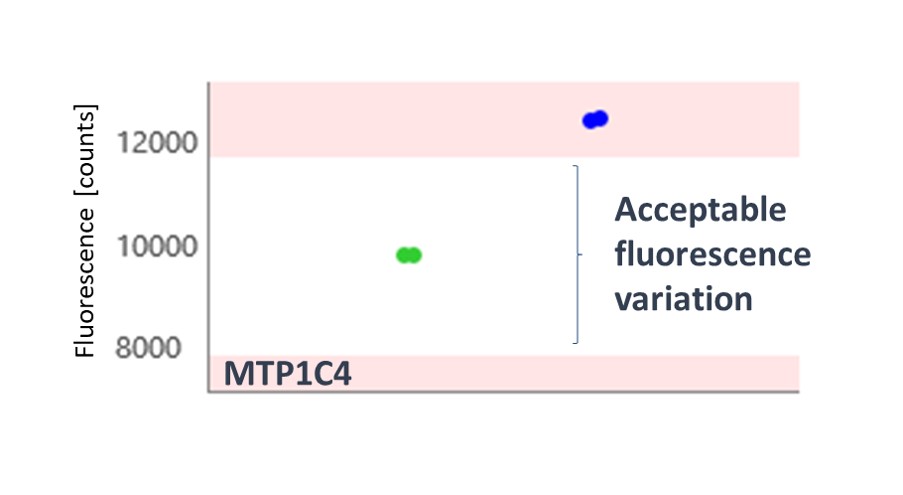
The thermophoresis traces allow easy identification of false-positive fragments such as aggregators and compounds effecting the fluorescence signal (Figures 3 and 4, respectively).
A 5.3% hit rate was obtained. Screening the same fragments using Differential Scanning Fluorimetry (DSF) or using an activity-based AlphaScreen assay both showed only a 0.3% hit rate. This illustrates the point that orthogonal biophysical methods can give different results in fragment screening, and it is important to find the best method for any given target. We find MST gives better results than most other biophysical methods, and because of its speed and low protein consumption, it is usually the screening approach that we try first.
Hits were taken into secondary screening to determine their binding affinities (Kd) to the G9a-SAM complex using MST (Examples shown in Table 1).
| Frag ID | Kd[μM] | LE | STD-NMR Positive Binding | X-Ray Structure Resolution |
|---|---|---|---|---|
| MTP3B6 | 19 | 0.66 | ✓ | 1.5 Å |
| MTP2C3 | 56 | 0.41 | X | |
| MTP4E1 | 115 | 0.41 | ✓ | X |
| MTP2D8 | 327 | 0.54 | ✓ | 2.0 Å |
| MTP3G10 | 411 | 0.53 | ✓ | 1.8 Å |
| MTP2H9 | 564 | 0.44 | ✓ | X |
| MTP3G1 | 718 | 0.36 | X |
Fragment hits were identified with good affinities, and hence excellent ligand efficiency (LE; a value of 0.3 is typically considered to be good).
Orthogonal Hit Validation
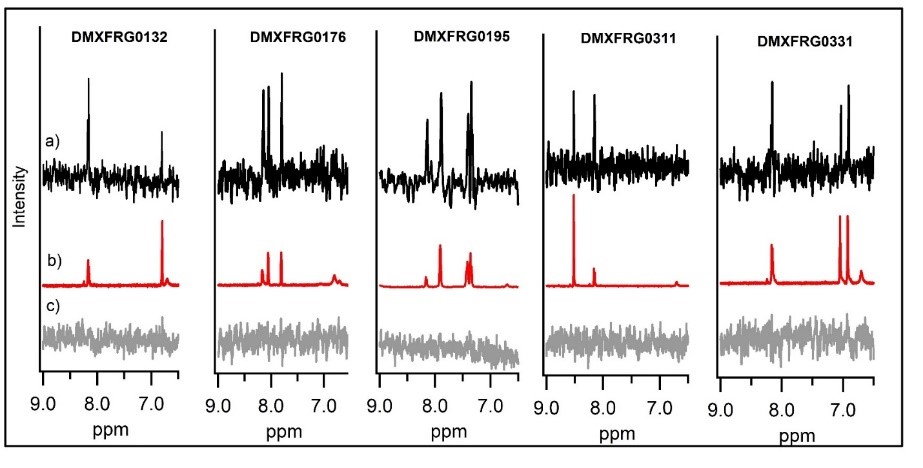
Orthogonal confirmation of hit binding to G9a was demonstrated by Saturation Transfer Difference (STD) NMR spectroscopy (Figure 5). Three G9a-fragment structures were solved in-house in the presence of the co-factor, SAM, with a resolution of 1.5–2.0 Å (Figure 6), which revealed different fragment binding modes. This resulted in several different options for fragment elaboration.
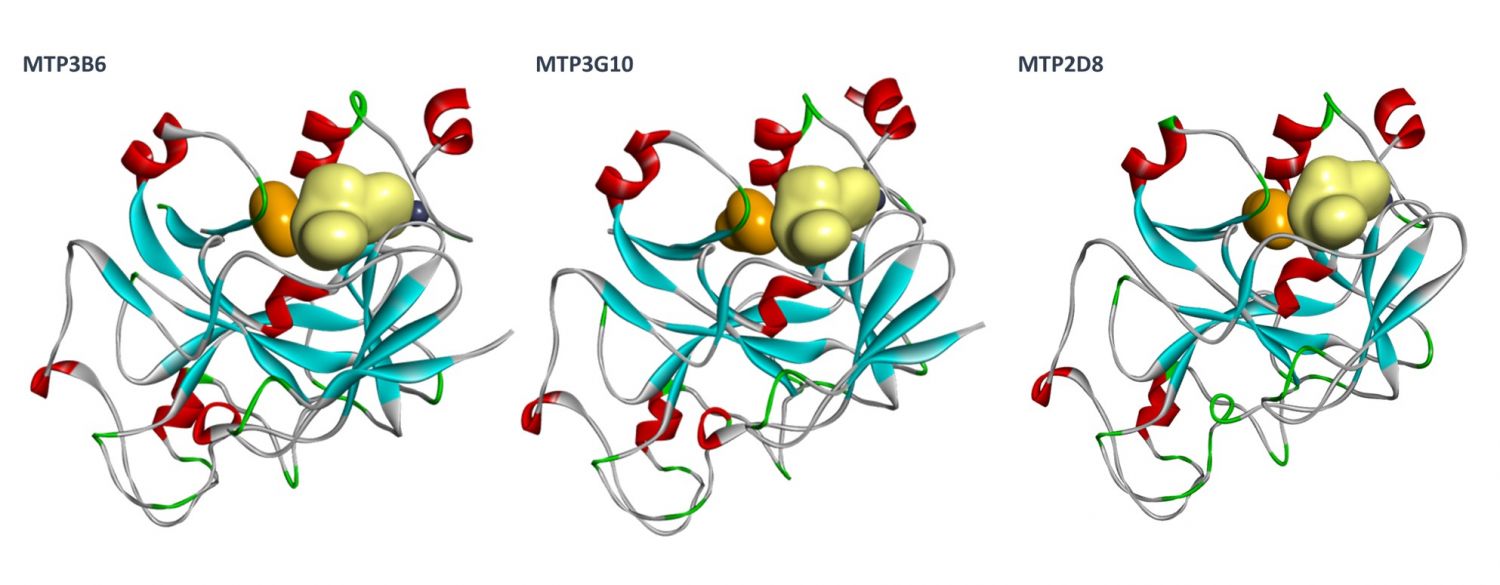
Fragment Elaboration
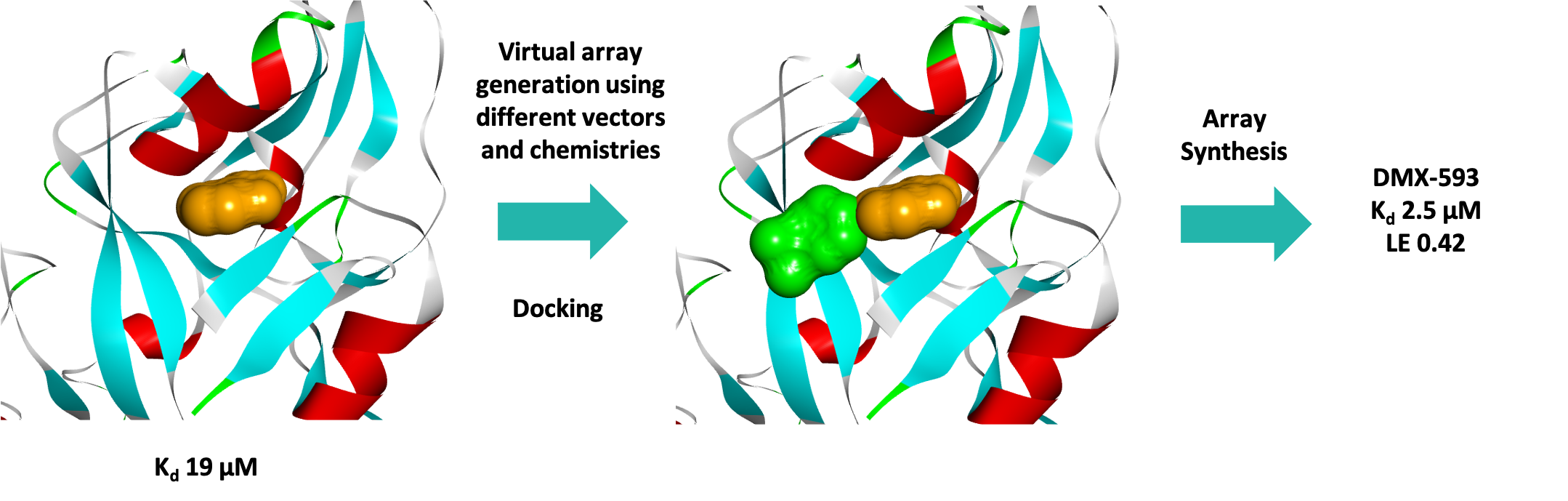
Three series were progressed and after one round of fragment elaboration (~50 compounds) a 10-fold increase in affinity was achieved. Figure 7 shows the efficient process used by the medicinal and computational chemists.
Conclusions
MST was used to successfully screen a sub-set of our fragment library against the KMT G9a with a hit rate of 5.3%, identifying several fragments with high ligand efficiencies. Three fragment hits were successfully crystallised bound to G9a, confirming that they were substrate rather than co-factor competitive inhibitors as was desired, and the structural data enabled a SBDD programme for this target. In just one round of fragment elaboration a 10-fold increase in affinity was achieved.
Domainex Expertise
• Integrated Drug Discovery • Fragment Screening • Hit Identification • MicroScale Thermophoresis (MST) Services
• Biophysics • Oncology • Medicinal and Synthetic Chemistry • Fragment-baed Drug Design (FBDD)
• X-ray Crystallography • Structural Biology • Structure-based Drug Design (SBDD) • Methyltransferase Drug Discovery
References
1. Deregulation of histone lysine methyltransferases contributes to oncogenic transformation of human bronchoepithelial cells. Hideo Watanabe, Kenzo Soejima, Hiroyuki Yasuda, Ichiro Kawada,Ichiro Nakachi, Satoshi Yoda, Katsuhiko Naoki and Akitoshi Ishizaka. Cancer Cell Int., 2008, 8, 15
2. Downregulation of histone H3 lysine 9 methyltransferase G9a induces centrosome disruption and chromosome instability in cancer cells. Yutaka Kondo, Lanlan Shen, Saira Ahmed, Yanis Boumber, Yoshitaka Sekido, Bassem R. Haddad and Jean-Pierre J. Issa. PLoS One, 2008, 3, 4
3. G9a mediates hypoxia-dependent gene repression. Francesco Casciello, Fares Al-Ejeh, Greg Kelly, Donal J. Brennan, Shin Foong Ngiow, Arabella Young, Thomas Stoll, Karolina Windloch, Michelle M. Hill, Mark J. Smyth, Frank Gannon, Jason S. Lee. PNAS, 2017, 114, (27), 7077-7082
4. Targeting histone methyltransferase G9a inhibits growth and Wnt signaling pathway by epigenetically regulating HP1α and APC2 gene expression in non-small cell lung cancer. Keqiang Zhang, Jinhui Wang, Lu Yang, Yate-Ching Yuan, Tommy R. Tong, Jun Wu, Xinwei Yun, Melissa Bonner, Rajendra Pangeni, Zheng Liu, Tiger Yuchi, Jae Y. Kim and Dan J. Raz. Molecular Cancer, 2018, 17, 153
5. MYC Interacts with the G9a Histone Methyltransferase to Drive Transcriptional Repression and Tumorigenesis. William B. Tu, Yu-Jia Shiah, Corey Lourenco, Peter J. Mullen, Dharmendra Dingar, Cornelia Redel, Aaliya Tamachi, Wail Ba-Alawi, Ahmed Aman, Rima Al-awar, David W. Cescon, Benjamin Haibe-Kains, Cheryl H. Arrowsmith, Brian Raught, Paul C. Boutros and Linda Z. Penn. Cancer Cell, 2018, 34, 4, 579-595
Start your next project with Domainex
Contact one of our experts today